Search Count: 22
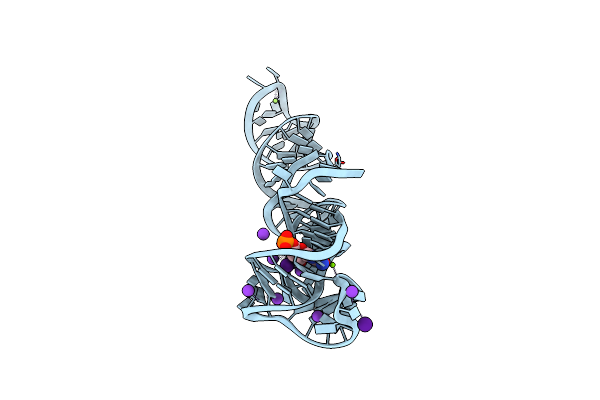 |
Cocrystal Structure Of Clostridium Beijerinckii Ztp Riboswitch With Zmp And Cs
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2024-11-27 Classification: RNA Ligands: CS, MG, AMZ, K |
 |
Co-Crystal Structure Of The Fusobacterium Ulcerans Ztp Riboswitch Using An X-Ray Free-Electron Laser
Organism: Fusobacterium ulcerans
Method: X-RAY DIFFRACTION Resolution:4.10 Å Release Date: 2019-07-17 Classification: RNA Ligands: MG, AMZ, K, CS |
 |
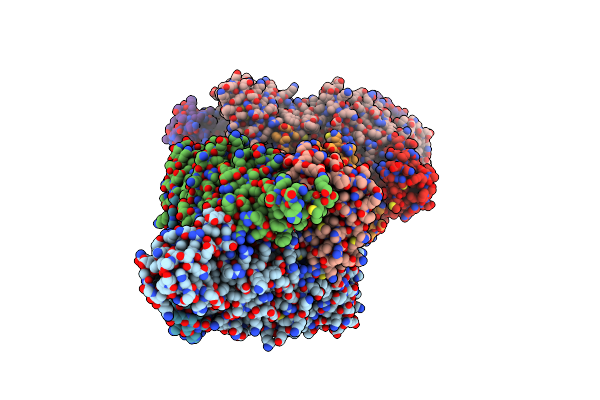
Human Liver Fructose-1,6-Bisphosphatase 1 (Fructose 1,6-Bisphosphate 1-Phosphatase, E.C.3.1.3.11) Complexed With The Allosteric Inhibitor 1-(3-Chlorophenyl)Sulfonyl-3-[5-[(3-Chlorophenyl)Sulfonylcarbamoylamino]Pentyl]Urea
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-09 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: 94J, AMZ |
 |
Crystal Structure Of Neanderthal Adenylosuccinate Lyase (Adsl)In Complex With Its Products Aicar And Fumarate
Organism: Homo sapiens neanderthalensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-05-30 Classification: LYASE Ligands: SSS, CL, FUM, AMZ |
 |
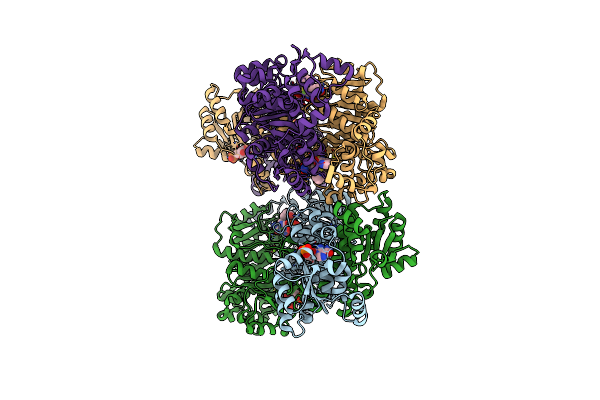
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2018-01-10 Classification: TRANSFERASE, HYDROLASE Ligands: 8UM, AMZ, MG |
 |
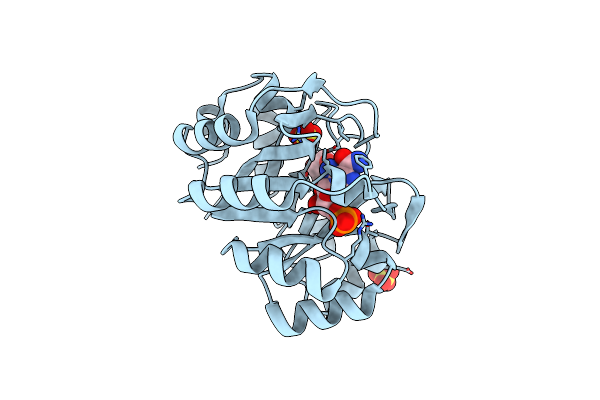
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2018-01-10 Classification: TRANSFERASE, HYDROLASE Ligands: AMZ, MG, 8US |
 |
Crystal Structure Of Phosphoribosyl Isomerase A From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-09-30 Classification: ISOMERASE Ligands: SO4, GOL, AMZ |
 |
Organism: Actinomyces odontolyticus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-09-09 Classification: RNA Ligands: AMZ, MG, IRI |
 |
Organism: Actinomyces odontolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-09-09 Classification: RNA Ligands: AMZ, MG, IRI, CAC |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2015-08-26 Classification: RNA Ligands: AMZ, MG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2015-08-12 Classification: RNA Ligands: MG, AMZ, K |
 |
Organism: Streptomyces sviceus atcc 29083
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-08-06 Classification: ISOMERASE Ligands: SO4, AMZ |
 |
Crystal Structure Of Mycobacterium Tuberculosis Purh Complexed With Aicar And A Novel Nucleotide Cfair, At 2.48 A Resolution.
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2011-09-28 Classification: TRANSFERASE-HYDROLASE Ligands: K, AMZ, PO4, JLN |
 |
Crystal Structure Of Faicar Synthetase (Purp) From M. Jannaschii Complexed With Amppcp And Aicar
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-12-04 Classification: LIGASE Ligands: SO4, CL, ACP, AMZ |
 |
Crystal Structure Of Faicar Synthetase (Purp) From M. Jannaschii Complexed With Atp And Aicar
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-12-04 Classification: LIGASE Ligands: SO4, CL, ATP, AMZ |
 |
Crystal Structure Of The Adenylate Sensor From Amp-Activated Protein Kinase In Complex With 5-Aminoimidazole-4-Carboxamide 1-Beta-D-Ribofuranotide (Zmp)
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2007-10-23 Classification: TRANSFERASE Ligands: AMZ |
 |
Crystal Structure Of A Cbs Domain Pair From The Regulatory Gamma1 Subunit Of Human Ampk In Complex With Amp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2007-05-08 Classification: TRANSFERASE Ligands: AMZ |
 |
Crystal Structure Of Puro/Aicar From Methanothermobacter Thermoautotrophicus
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-04-24 Classification: HYDROLASE Ligands: AMZ |
 |
Atomic Resolution Structure Of Saicar-Synthase From Saccharomyces Cerevisiae Complexed With Adp, Aicar, Succinate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2006-06-08 Classification: LIGASE Ligands: ADP, AMZ, AMP, SIN, MG, SO4 |
 |
Crystal Structure Of Human Atic In Complex With Folate-Based Inhibitor, Bw2315U89Uc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2004-04-27 Classification: TRANSFERASE, HYDROLASE Ligands: K, XMP, AMZ, BW2 |

