Search Count: 8
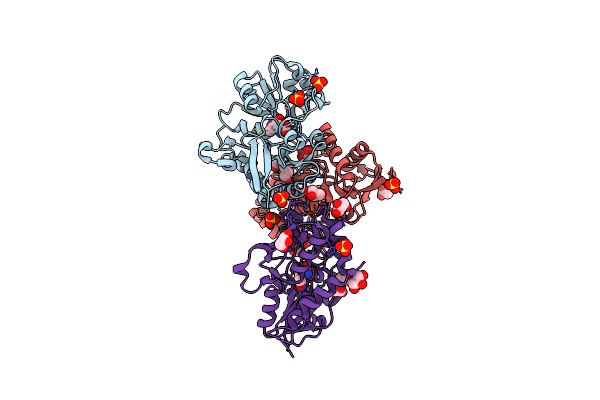 |
X-Ray Structure Of Iglur4 Flip Ligand-Binding Core (S1S2) In Complex With (S)-Ampa At 1.90A Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-12-09 Classification: MEMBRANE PROTEIN Ligands: AMQ, SO4, GOL, ACY |
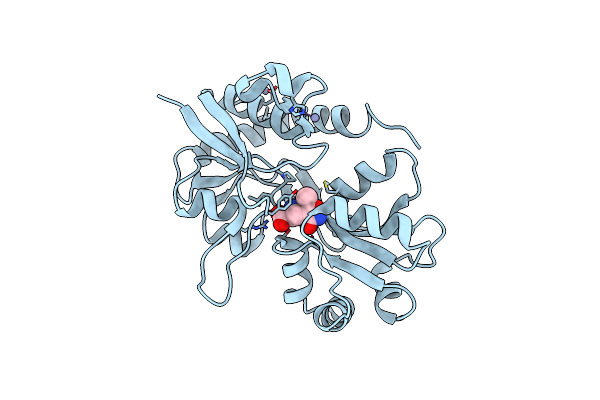 |
Crystal Structure Of The Binding Domain Of The Ampa Subunit Glur3 Bound To Ampa
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2008-11-25 Classification: SIGNALING PROTEIN Ligands: ZN, AMQ |
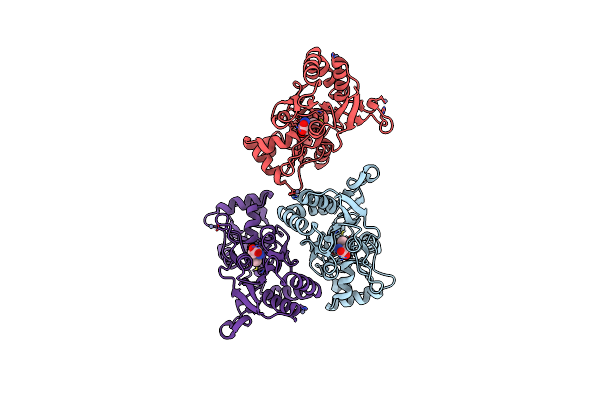 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-06-10 Classification: MEMBRANE PROTEIN Ligands: ZN, AMQ |
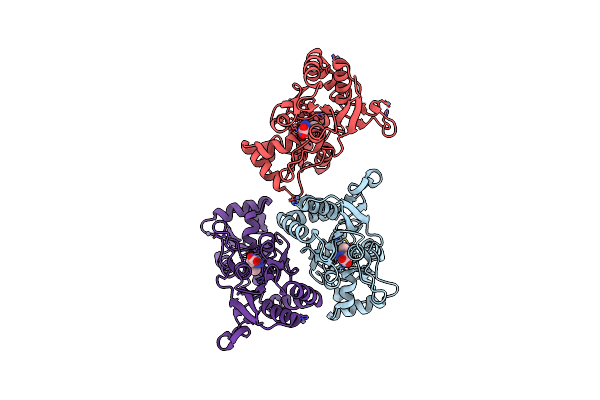 |
Crystal Structure Of The Glur2 Ligand Binding Core (S1S2J) L650T Mutant In Complex With Ampa
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-06-10 Classification: MEMBRANE PROTEIN Ligands: ZN, AMQ |
 |
Crystal Structure Of The Glur2 Ligand-Binding Core (S1S2J) L650T Mutant In Complex With Ampa (Ammonium Sulfate Crystal Form)
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-06-10 Classification: MEMBRANE PROTEIN Ligands: SO4, AMQ |
 |
Crystal Structure Of The Glur2 Ligand-Binding Core (S1S2J) With The L483Y And L650T Mutations And In Complex With Ampa
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-06-10 Classification: MEMBRANE PROTEIN Ligands: SO4, AMQ |
 |
Crystal Structure Of The Non-Desensitizing Glur2 Ligand Binding Core Mutant (S1S2J-L483Y) In Complex With Ampa At 2.3 Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-06-05 Classification: MEMBRANE PROTEIN Ligands: AMQ |
 |
Crystal Structure Of The Glur2 Ligand Binding Core (S1S2J) In Complex With Ampa At 1.7 Resolution
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2000-11-01 Classification: MEMBRANE PROTEIN Ligands: ZN, AMQ |

