Search Count: 3,262
All
Selected
 |
Crystal Structure Of The Daba Transaminase Ectb From The Halophilic And Cold-Adapted Marinobacter Sp. Ck1 -Mutant K264A
Organism: Marinobacter sp. ck-1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2026-02-18 Classification: TRANSFERASE Ligands: PLP, EDO, SO4, NA |
 |
Crystal Structure Of The Daba Transaminase Ectb From The Halophilic And Cold-Adapted Marinobacter Sp. Ck1
Organism: Marinobacter sp. ck-1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-02-18 Classification: TRANSFERASE Ligands: PMP, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: ITN |
 |
Crystal Structure Of A Transaminase Pata From Pseudonocardia Ammonioxydans In Complex With Plp And Llp
Organism: Pseudonocardia ammonioxydans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PLP |
 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-11-19 Classification: TRANSFERASE |
 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: P7I |
 |
Structure Of The K193M Mutant Of Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: PLP, P7I, CL |
 |
Human Mitochondrial Aspartate Aminotransferase Complex With Plp At 1.5 Angstrom.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: GOL, SO4, CL |
 |
Organism: Streptomyces sp. bv333
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2025-09-10 Classification: TRANSFERASE Ligands: PXG, EDO, PEG, CA, CL |
 |
Organism: Streptomyces sp. bv333
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-08-27 Classification: TRANSFERASE Ligands: EDO, PLP, SO4 |
 |
C387S Variant Of D-Ornithine/D-Lysine Decarboxylase Complexed With Pmp And 4-Guanidinobutanal
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2025-08-06 Classification: LYASE Ligands: DMS, NA, PMP, A1AZK, ACT, CL |
 |
Structure Of Tf9 Mutant Of Aminotransferase From Mycolicibacterium Neoaurum In Complex With Llp And G4O
Organism: Mycolicibacterium neoaurum vkm ac-1815d
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: G4O |
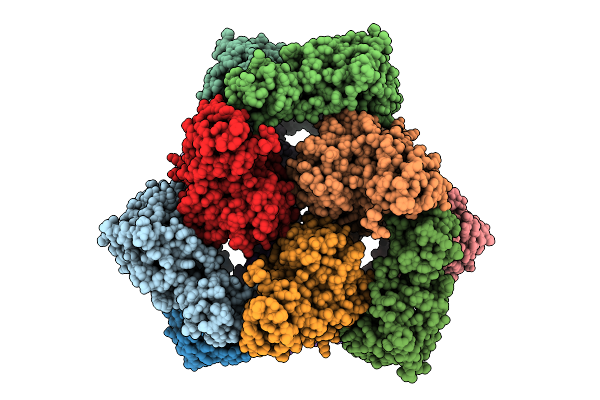 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSFERASE |
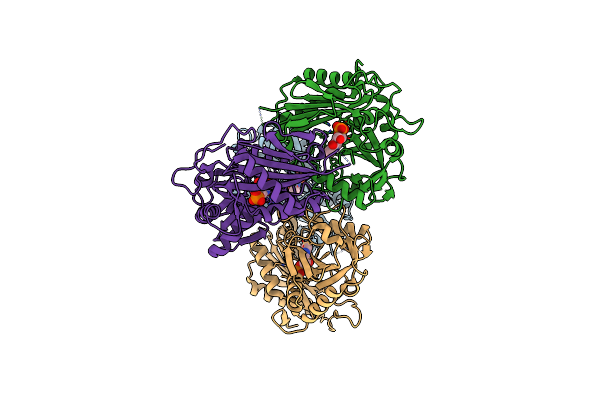 |
Structure Of Aminotransferase From Mycolicibacterium Neoaurum In Complex With Plp
Organism: Mycolicibacterium neoaurum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-06-18 Classification: TRANSFERASE Ligands: PLP |
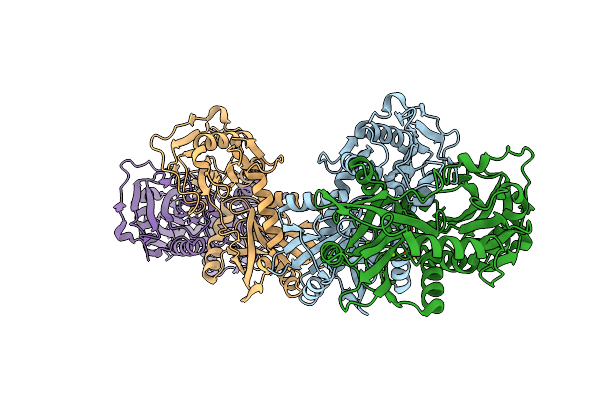 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-06-11 Classification: TRANSFERASE Ligands: MG |
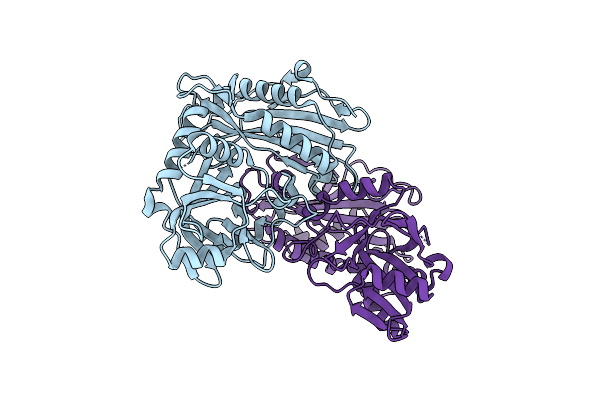 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2025-06-11 Classification: TRANSFERASE |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: PROTEIN TRANSPORT |
 |
Crystal Structure Of Omega Transaminase Ta_2799 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: GOL, EDO, PEG |
 |
Crystal Structure Of The L322F Mutant Of Omega Transaminase Ta_2799 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: PEG, EDO, SO4, GOL |
 |
Crystal Structure Of The Open State Of Omega Transaminase Ta_5182 From Pseudomonas Putida Kt2440
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2025-06-04 Classification: TRANSFERASE Ligands: SO4 |

