Search Count: 2,270
All
Selected
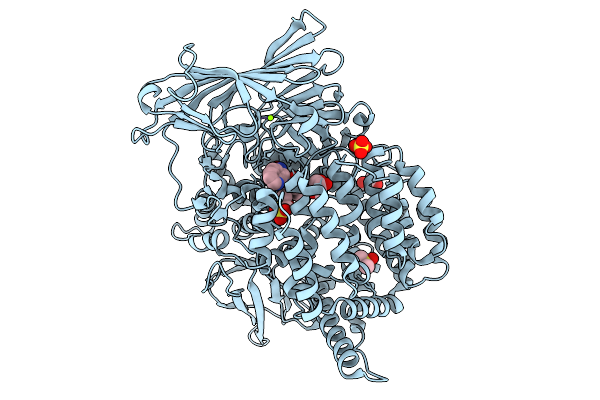 |
Plasmodium Falciparum M1 Aminopeptidase (Pfa-M1) Bound To Inhibitor 3K (Mips3415)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: DMS, A1CTH, GOL, ZN, MG, SO4 |
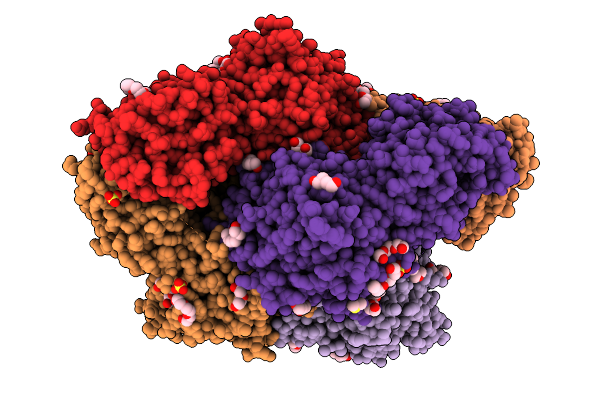 |
Plasmodium Falciparum M17 Aminopeptidase (Pfa-M17) Bound To Inhibitor 3Ab (Mips3413)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: ZN, CO3, PEG, SO4, A1CT4, 1PE, DMS, 2PE, A1CT5 |
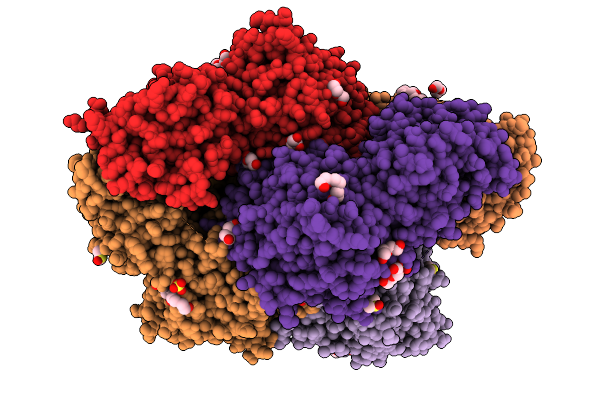 |
Plasmodium Falciparum M17 Aminopeptidase (Pfa-M17) Bound To Inhibitor 3K (Mips3415)
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: ZN, CO3, PEG, DMS, SO4, GOL, A1CTH, 1PE |
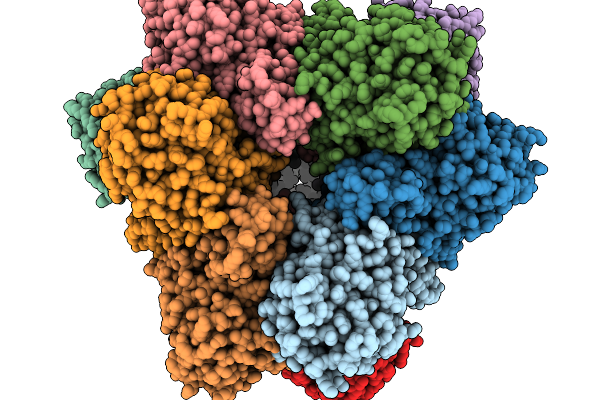 |
Organism: Methanocaldococcus jannaschii
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: CYTOSOLIC PROTEIN |
 |
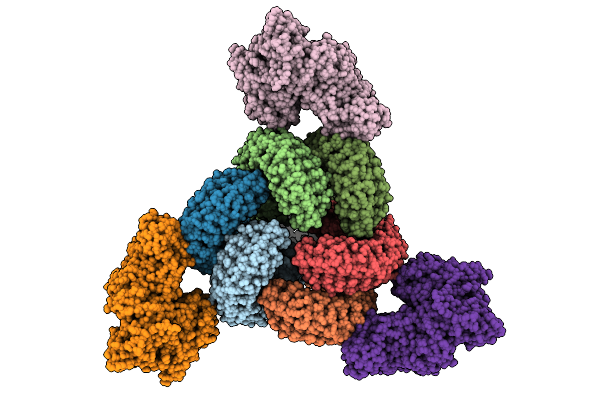
Organism: Feline infectious peritonitis virus (strain 79-1146), Felis catus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
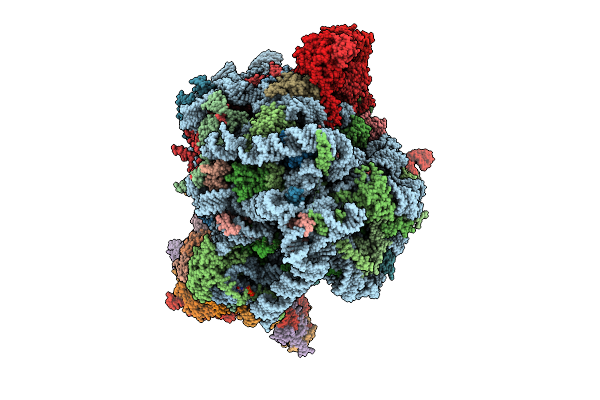 |
Human Quaternary Complex Of A Translating 80S Ribosome, Nac, Metap1 And Natd
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Aequorea victoria, Brachypodium distachyon
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: ZN, COA, GTP |
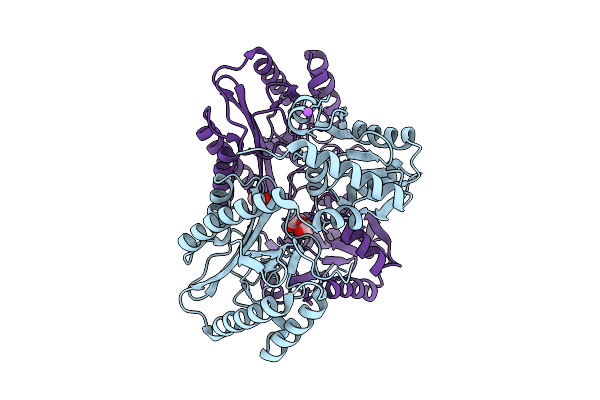 |
Crystal Structure Of Proline Dipeptidase From Pyrococcus Furiosus Complexed With Co
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-12-10 Classification: HYDROLASE |
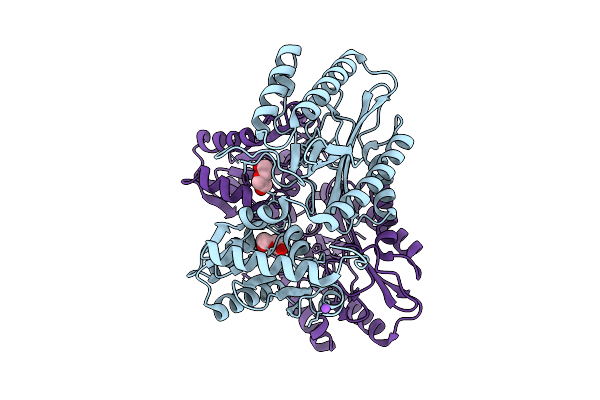 |
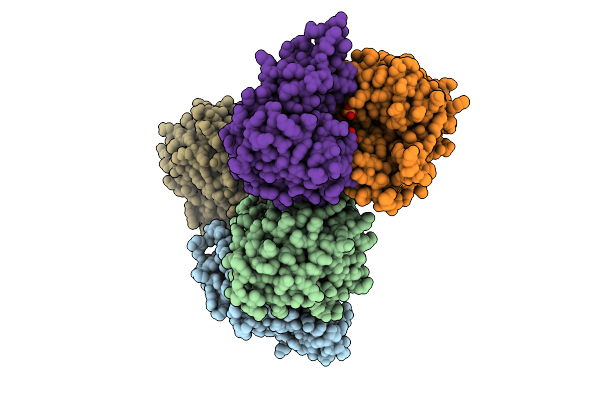
Crystal Structure Of Proline Dipeptidase From Pyrococcus Furiosus Complexed With Mn
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MN, NA, PEG |
 |
Crystal Structure Of Ttl[Nle], Thermophilic Lipase Ttl From Thermoanaerobacter Thermohydrosulfuricus Containing Non-Canonical Amino Acid Nle At The Position Of Met
Organism: Thermoanaerobacter thermohydrosulfuricus
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2025-10-29 Classification: LIPID BINDING PROTEIN Ligands: SO4 |
 |
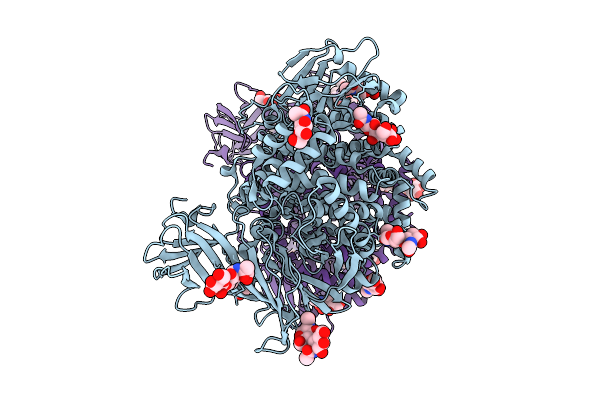
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Resolution:3.87 Å Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: ZN, ARG |
 |
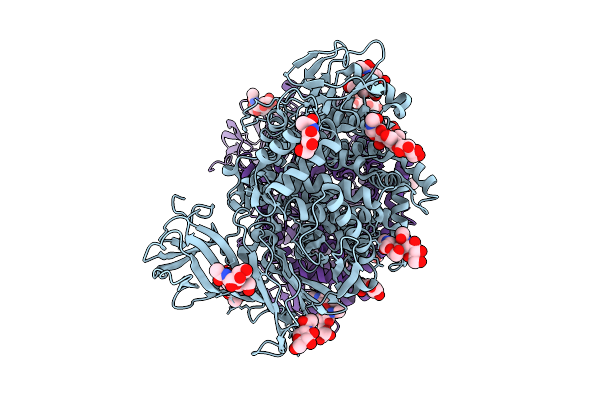
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: HYDROLASE Ligands: NAG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN Ligands: NAG, ZN |
 |
Molecular Basis Of Pathogenicity Of The Recently Emerged Fcov-23 Coronavirus. Complex Of Fapn With Fcov-23 Rbd
Organism: Felis catus, Feline coronavirus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG, ZN |
 |
Cryo-Em Map Of 30S Ribosomal Subunit In Complex With Metap1C Of Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RIBOSOME |
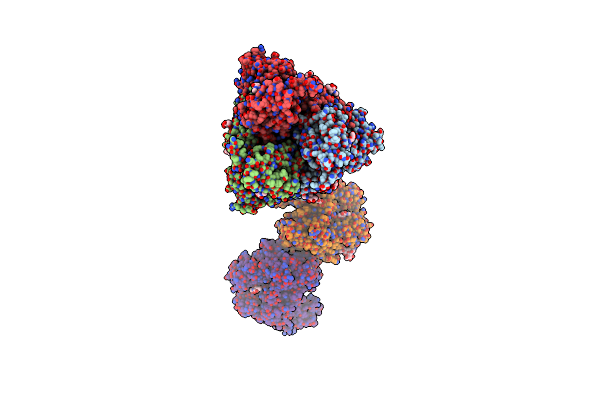 |
Cryoem Structure Of The Spike Protein Of Human Cov 229E In Complex With Receptor Hapn (Composite Map)
Organism: Human coronavirus 229e, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Erap1 In Complex With 1-[2-(3-Oxo-3,4-Dihydro-2H-1,4-Benzothiazin-4-Yl)Acetamido]Cyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, B3P, EDO, BR, A1IMK |
 |
Erap1 In Complex With 1-[2-(6-Bromo-3-Oxo-3,4-Dihydro-2H-1,4-Benzoxazin-4-Yl)Acetamido]-4,4-Difluorocyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, PO4, EDO, A1IMJ |
 |
Erap1 In Complex With 1-[2-(6-Chloro-3-Oxo-3,4-Dihydro-2H-1,4-Benzothiazin-4-Yl)Acetamido]Cyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, MLT, EDO, A1IMM |
 |
Erap1 In Complex With 1-[2-(2-Oxo-5-Phenyl-2,3-Dihydro-1,3-Benzothiazol-3-Yl)Acetamido]Cyclohexane-1-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2025-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN, PO4, EDO, A1IML |

