Search Count: 710
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: ACY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: PTR, SO4 |
 |
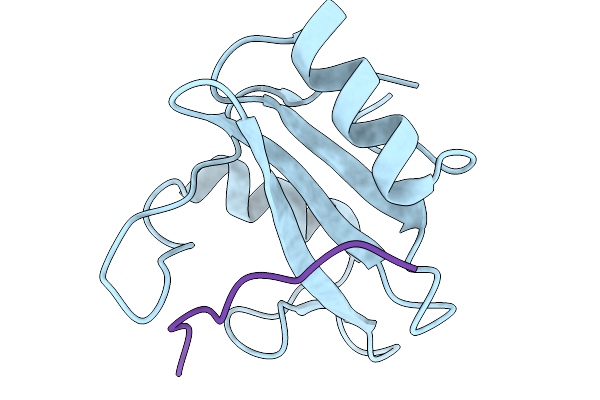
Crystal Structure Of N-Sh2 Domain Of Shp2 Bound To Gab1 Tyrosine Phosphorylated Peptide (624-633) Qvepyldldld
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: PO4 |
 |
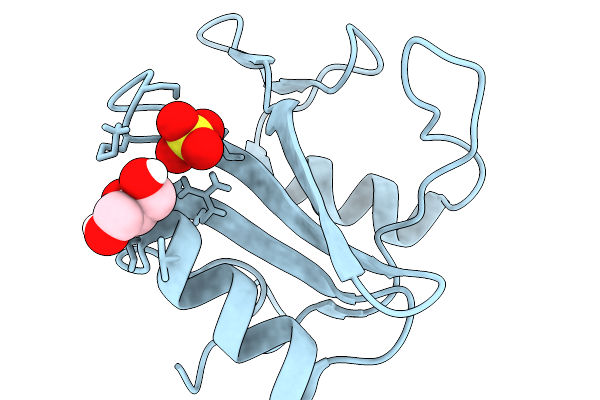
Crystal Structure Of N-Sh2 Domain Of Shp2 With T42A Mutation Bound To Gab1 Tyrosine Phosphorylated Peptide (624-633) Qvepyldldld
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING |
 |
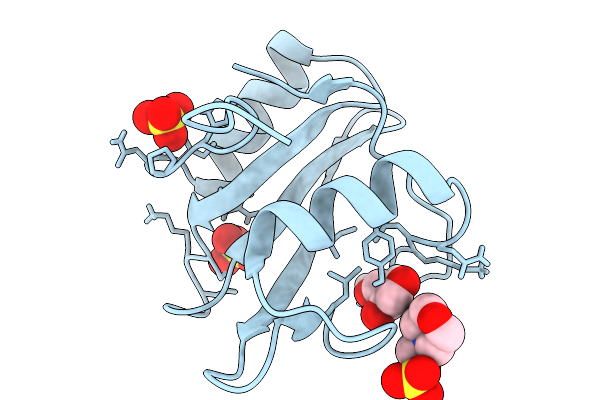
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: GOL, SO4 |
 |
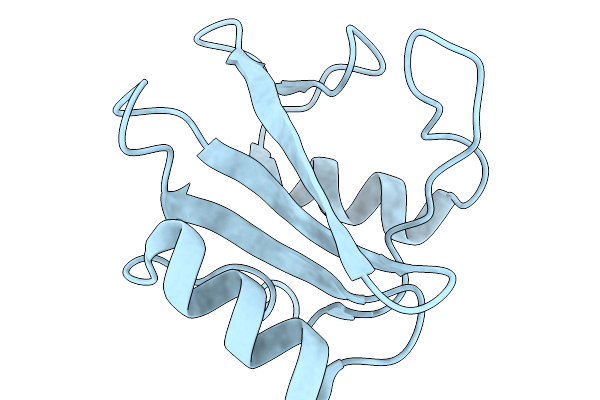
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING Ligands: MES, GOL, SO4 |
 |
Crystal Structure Of Shorter Construct Of Shp2 Unbound N-Sh2 Domain (Y66 In Blocking Conformation)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: PROTEIN BINDING |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: STRUCTURAL PROTEIN Ligands: NAP, ACO, MG |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: STRUCTURAL PROTEIN Ligands: NAP, MG |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: STRUCTURAL PROTEIN Ligands: NAP, MG |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN Ligands: NAP, MG |
 |
Organism: Bdellovibrio bacteriovorus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN Ligands: NAP |
 |
Organism: Bdellovibrio bacteriovorus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN Ligands: ACO, NAP |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN Ligands: MG |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN Ligands: SO4 |
 |
Organism: Murine norovirus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: VIRUS Ligands: CA |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, TPP, DNA, MG, CL |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2025-04-16 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, CL, TPP, DNA, EDO, MG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: LYASE Ligands: GOL, PGE, EPE |

