Search Count: 310
All
Selected
 |
The Cryo-Em Structure Of Human Tissue Nonspecific Alkaline Phosphatase (Htnap) In Complex With Mls-0038949
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: HYDROLASE Ligands: MG, ZN, CA, A1JNY |
 |
Structure Of Phosphonate Monoester Hydrolase From Rhizobium Leguminosarum With Vanadate
Organism: Rhizobium johnstonii 3841
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MN, VO4, NA |
 |
Structure Of Phosphonate Monoester Hydrolase From Rhizobium Leguminosarum With Phenyl Phosphonate
Organism: Rhizobium johnstonii 3841
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-10-08 Classification: HYDROLASE Ligands: MN, SV7, ACT, NA |
 |
The Crystal Structure Of Human Tissue Nonspecific Alkaline Phosphatase (Htnap) In Complex With Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2025-09-17 Classification: HYDROLASE Ligands: ZN, MG, CA, NAG, PO4 |
 |
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: BR, CA, ACT |
 |
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: SO4, MG |
 |
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: CA, ACT, BEF |
 |
Organism: Flavobacterium johnsoniae uw101
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: ZN, GOL |
 |
Organism: Flavobacterium johnsoniae (strain atcc 17061 / dsm 2064 / jcm 8514 / nbrc 14942 / ncimb 11054 / uw101)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-04-17 Classification: SIGNALING PROTEIN Ligands: CA, GOL |
 |
The Cryo-Em Structure Of Human Tissue Nonspecific Alkaline Phosphatase And Single-Chain Fragment Variable (Scfv) Complex.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: HYDROLASE Ligands: NAG, MG, ZN, CA |
 |
The Crystal Structure Of Human Tissue Nonspecific Alkaline Phosphatase (Alpl) At Basic Ph
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: NAG, MG, ZN, CA |
 |
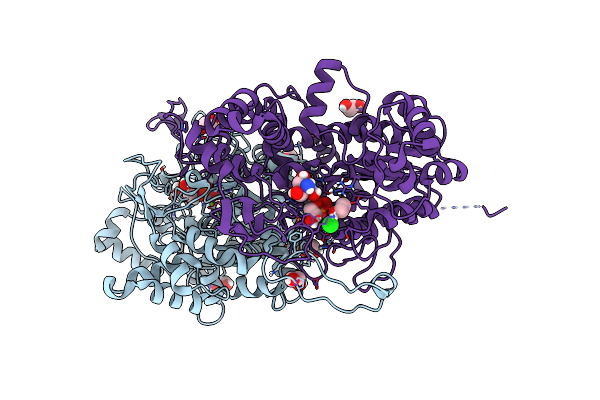
The Crystal Structure Of Human Tissue Nonspecific Alkaline Phosphatase (Alpl) At Acidic Ph
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2023-07-19 Classification: HYDROLASE Ligands: NAG, MG, ZN, CA |
 |
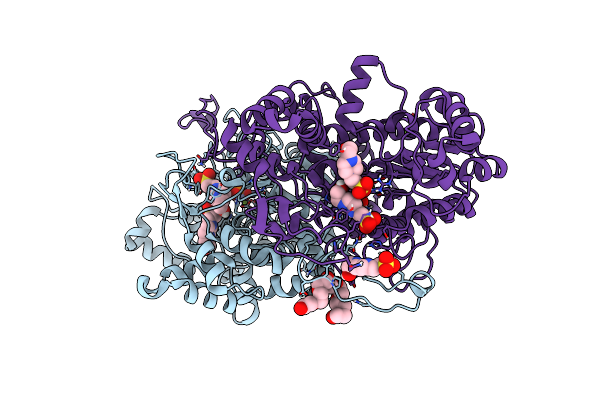
Organism: Vibrio sp. g15-21
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: ZN, MG, PO4, EDO, CL, TRS |
 |
Organism: Vibrio sp. g15-21
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: ZN, MG, 15P, CL, EPE, PO4 |
 |
Organism: Vibrio sp. g15-21
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: ZN, MG, EDO, PO4, CL |
 |
Organism: Vibrio sp. g15-21
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: ZN, BR, MG, PO4 |
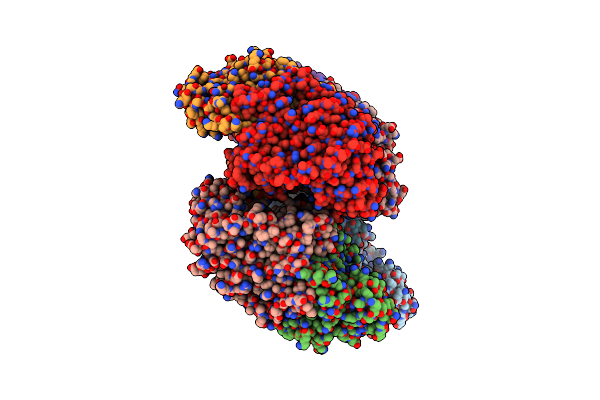 |
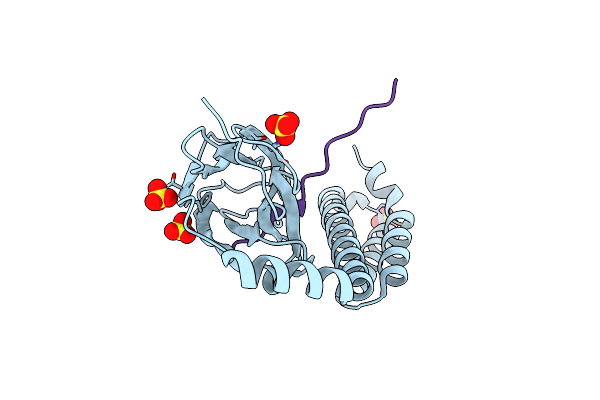
Crystal Structure Of Bacterial Alkaline Phosphatase From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae hs11286
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-02-02 Classification: HYDROLASE Ligands: ZN, MG |
 |
Crystal Structure Of The Substrate-Binding Domain Of E. Coli Dnak In Complex With The Peptide Rkqstialallpllftprr
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-06-23 Classification: CHAPERONE/HYDROLASE Ligands: SO4 |
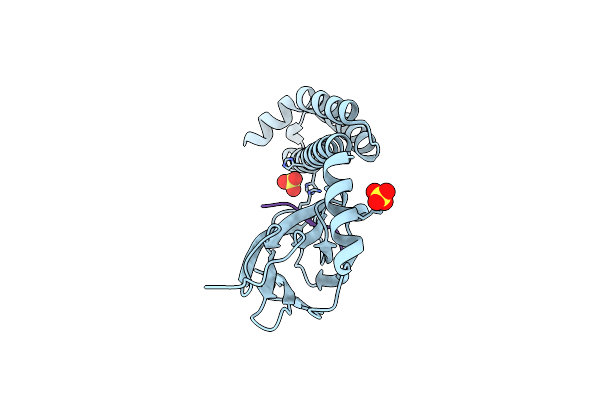 |
Crystal Structure Of The Substrate-Binding Domain Of E. Coli Dnak In Complex With The Peptide Ralallplsr
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-06-23 Classification: CHAPERONE/HYDROLASE Ligands: SO4 |
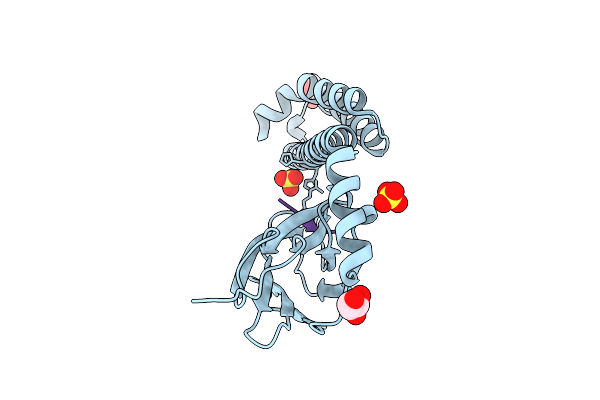 |
Crystal Structure Of The Substrate-Binding Domain Of E. Coli Dnak In Complex With The Peptide Eanqqkpllglfadg
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-06-23 Classification: CHAPERONE/HYDROLASE Ligands: GOL, SO4 |

