Search Count: 2,691
All
Selected
 |
Cryoem Structure Of Gfp-Like Protein From Aequorea Coerulescens With Trimbody
Organism: Synthetic construct, Thermotoga maritima msb8, Aequorea coerulescens
Method: ELECTRON MICROSCOPY Resolution:2.29 Å Release Date: 2026-02-11 Classification: STRUCTURAL PROTEIN |
 |
Organism: Synthetic construct, Thermotoga maritima msb8, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: STRUCTURAL PROTEIN |
 |
Organism: Synthetic construct, Thermotoga maritima msb8, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.43 Å Release Date: 2026-02-11 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Retro-Aldolase Ra95.5-8 Mutant With A Covalently Bound Cyclohexenone Derivative
Organism: Saccharolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2026-02-11 Classification: HYDROLASE Ligands: SO4, EDO |
 |
Crystal Structure Of Retro-Aldolase T53L/K210H Ra95.5-8 With A Covalently Bound Cyclohexenone Derivative
Organism: Saccharolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2026-02-11 Classification: HYDROLASE Ligands: SO4, EDO |
 |
Organism: Synthetic construct, Thermotoga maritima (strain atcc 43589 / dsm 3109 / jcm 10099 / nbrc 100826 / msb8), Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: STRUCTURAL PROTEIN |
 |
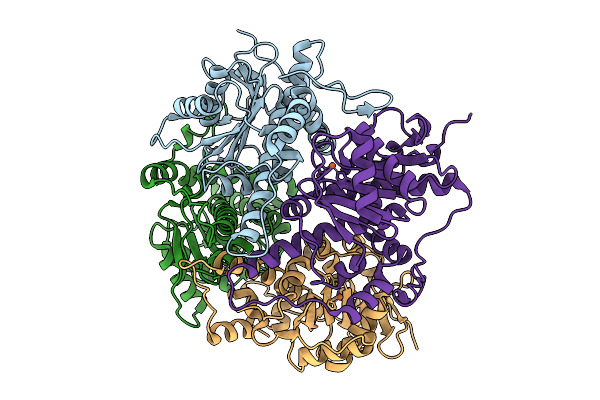
Crystal Structure Of None-Heme Iron Enzyme (Tqam) From Trichoderma Atroviride Bound With Iron And 2-Aminoisobutyric Acid
Organism: Trichoderma atroviride
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-02-04 Classification: METAL BINDING PROTEIN Ligands: FE, AIB |
 |
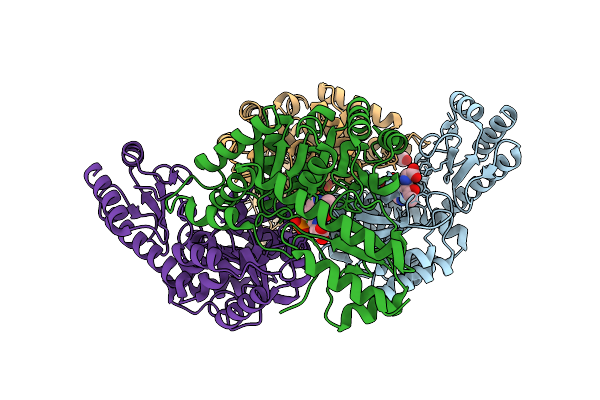
Crystal Structure Of None-Heme Iron Enzyme (Tqam) From Trichoderma Atroviride Bound With Iron
Organism: Trichoderma atroviride
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
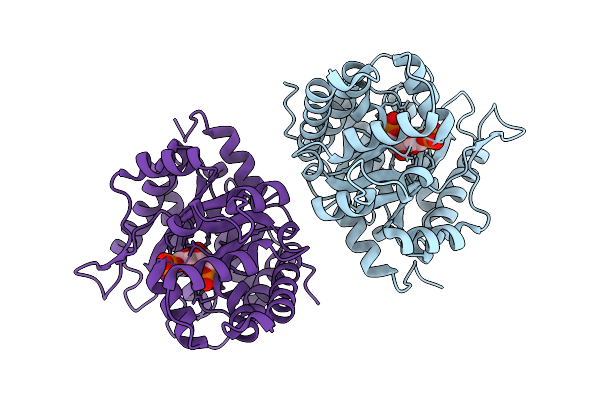
Crystal Structure Of L-Threonine Aldolase N18S/Q39R/Y319L Triple Mutant In Complex With Glycine From Neptunomonas Marina
Organism: Neptunomonas marina
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2026-01-21 Classification: LYASE Ligands: PLG, GLY |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2026-01-14 Classification: HYDROLASE Ligands: P6F |
 |
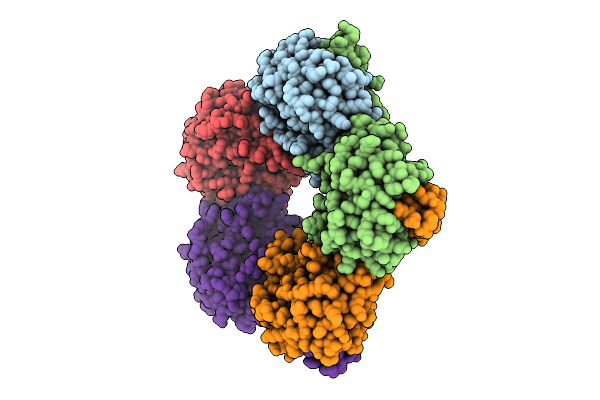
Crystal Structure Of L-Threonine Aldolase N18S/Q39R/Y319L Triple Mutant In Complex With Glycine From Neptunomonas Marina
Organism: Neptunomonas marina
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2026-01-07 Classification: LYASE Ligands: GLY, PLG |
 |
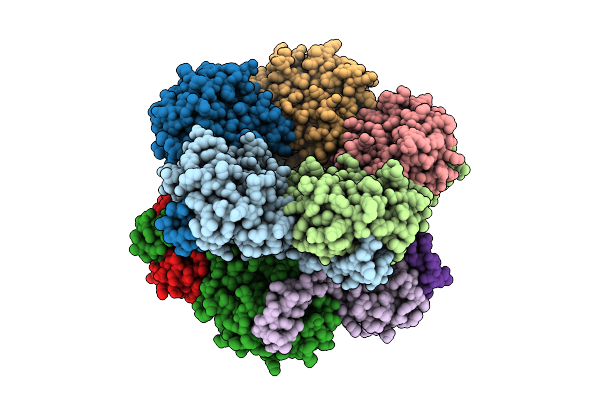
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Aeromonas sp. asnih1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
 |
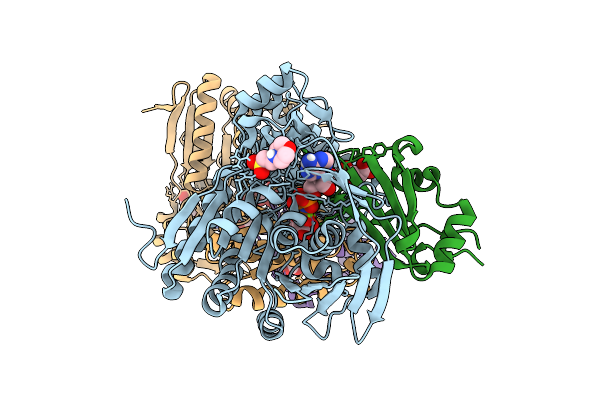
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Gilliamella apicola
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
 |
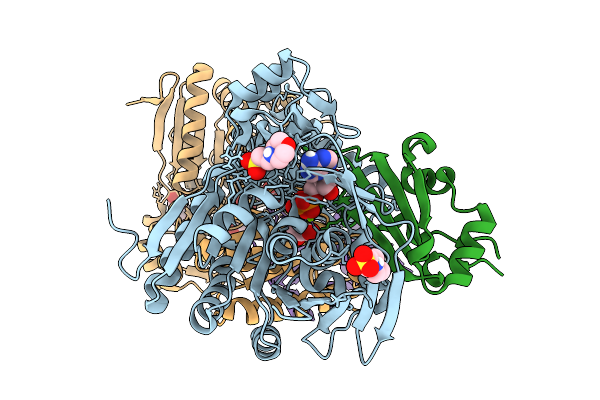
Organism: Klebsiella michiganensis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-10-22 Classification: LYASE Ligands: ANP, MG, A1H6S, MES, CL, S2G |
 |
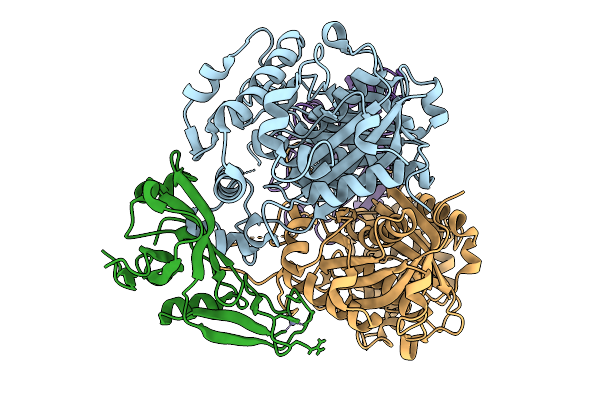
Organism: Raoultella planticola
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-10-08 Classification: LYASE Ligands: ADP, MG, PYR, MES, CL |
 |
Organism: Comamonas testosteroni kf-1
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-10-08 Classification: LIPID TRANSPORT Ligands: ZN |
 |
Crystal Structure Of The Complex Between Nb474 Mutant R53A,D125A And Trypanosoma Congolense Fructose-1,6-Bisphosphate Aldolase
Organism: Trypanosoma congolense il3000, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-10-01 Classification: LYASE Ligands: ACT, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2025-08-13 Classification: DE NOVO PROTEIN Ligands: ACT, GOL, NA |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2025-07-30 Classification: TRANSFERASE |
 |
X-Ray Structure Of Enterobacter Cloaca Transaldolase In Complex With D-Fructose-6-Phosphate.
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: F6R |

