Search Count: 15
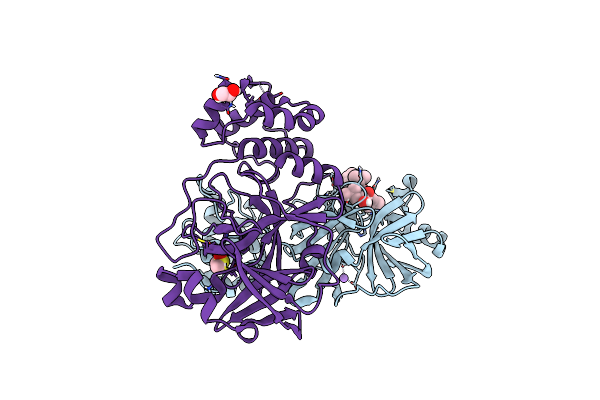 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, CL, NA, DMS, EDO |
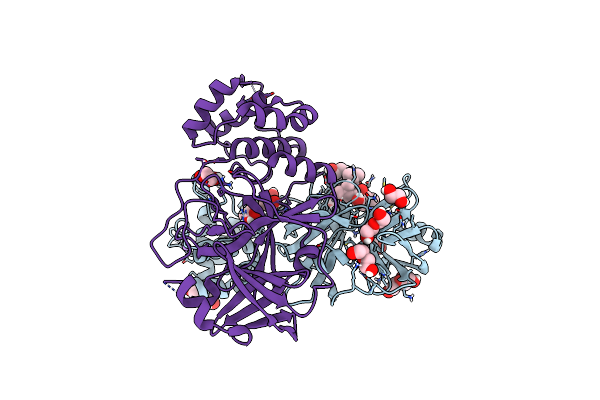 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Mm Mg-132, From An "Old" Crystal.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, EDO, PEG, NA, CL |
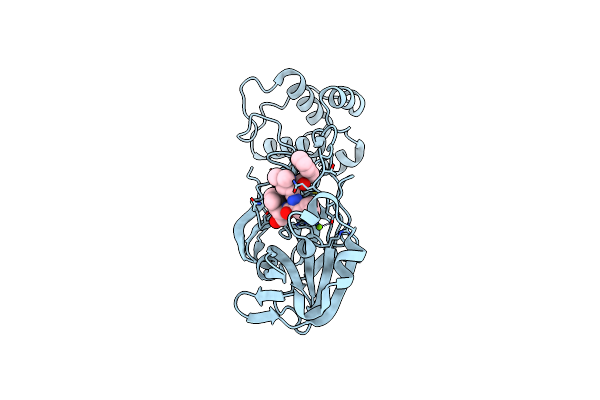 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-08-02 Classification: VIRAL PROTEIN Ligands: ALD, MG |
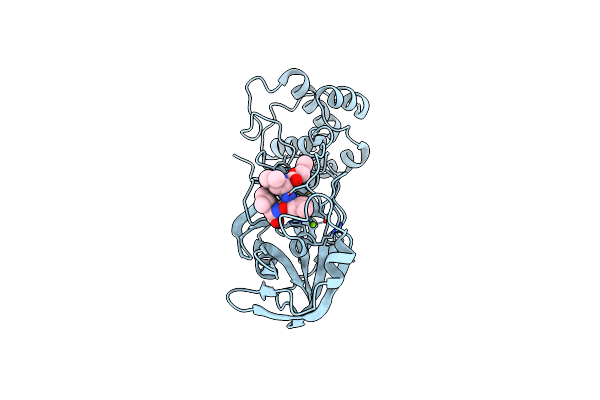 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-08-02 Classification: VIRAL PROTEIN Ligands: MG, ALD |
 |
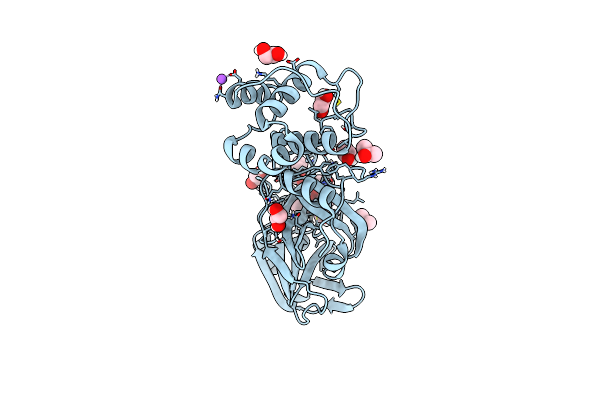
Crystal Structure Of Human Cathepsin L In Complex With Covalently Bound Mg132
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: ALD, PEG, ACT |
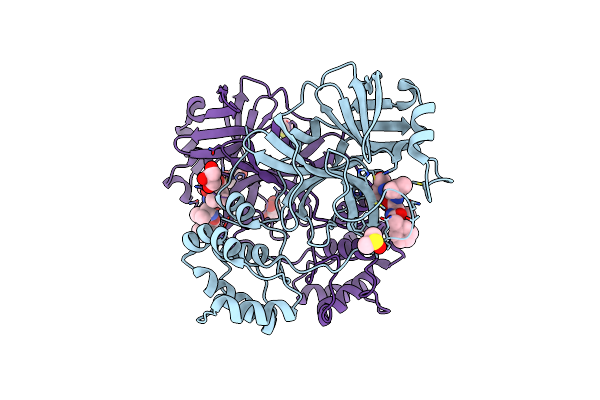 |
Crystal Structure Of The Sars-Cov-2 (Covid-19) Main Protease In Complex With Mg132
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2021-06-23 Classification: VIRAL PROTEIN Ligands: DMS, ALD, GOL |
 |
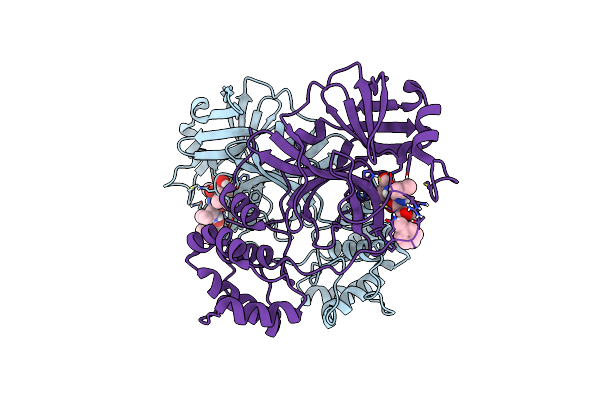
Crystal Structure Of Mg-132 Covalently Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 In Spacegroup C2.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-04-07 Classification: VIRAL PROTEIN Ligands: ALD, EDO, IPA, CL, NA |
 |
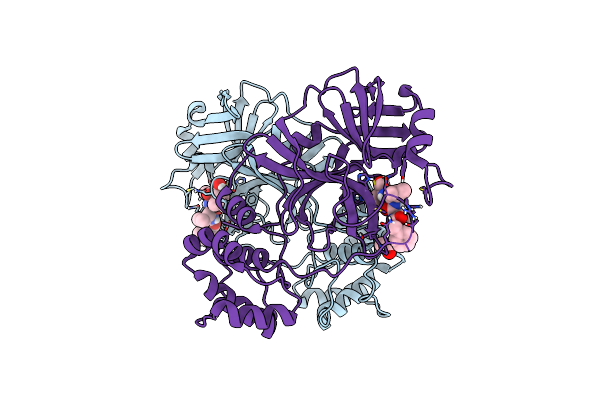
Crystal Structure Of Mg-132 Covalently Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 In Spacegroup P1.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-07 Classification: VIRAL PROTEIN Ligands: CL, ALD |
 |
Crystal Structure Of Mg-132 Covalently Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 In Spacegroup P1 In Absence Of Dtt.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2021-04-07 Classification: VIRAL PROTEIN Ligands: ALD, ACT, CL |
 |
Crystal Structure Of Mg-132 Covalently Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2021-03-03 Classification: VIRAL PROTEIN Ligands: ALD, EDO, PEG, NA, CL |
 |
Crystal Structure Of Mg-132 Covalently Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 In Absence Of Dtt.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2021-03-03 Classification: VIRAL PROTEIN Ligands: EDO, ALD, CL, NA |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-03-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MG, CL, ALD |
 |
Yeast 20S Proteasome Beta5-K33A Mutant (Propeptide Expressed In Trans) In Complex With Mg132
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-03-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MG, CL, ALD |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-02-12 Classification: HYDROLASE/HYDROLASE Inhibitor Ligands: MG, ALD |
 |
Use Of Papain As A Model For The Structure-Based Design Of Cathepsin K Inhibitors. Crystal Structures Of Two Papain Inhibitor Complexes Demonstrate Binding To S'-Subsites.
Organism: Carica papaya
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-08-12 Classification: HYDROLASE Ligands: ALD |

