Search Count: 187
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: BIOSYNTHETIC PROTEIN Ligands: ALA, ZN |
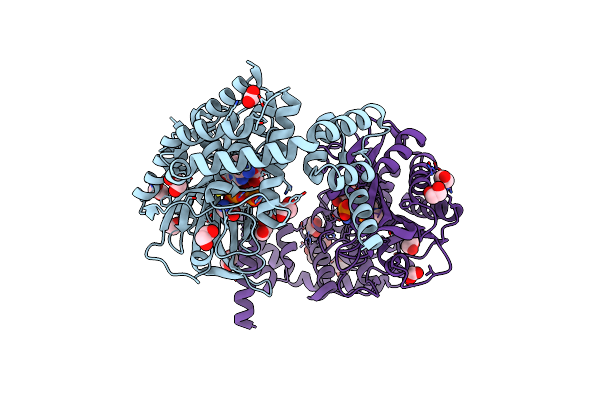 |
Crystal Structure Of Alanyl-Trna Synthetase In Complex With Atp And L-Alanine
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: ATP, ALA, EDO, FMT, MLT, CIT |
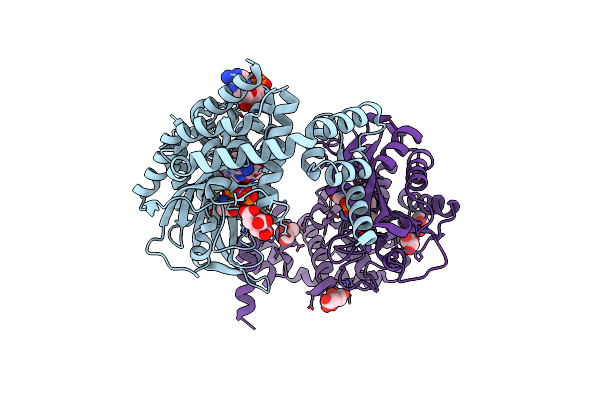 |
Crystal Structure Of Alanyl-Trna Synthetase L219M Mutant In Complex With Atp And L-Alanine
Organism: Methylomonas sp. dh-1
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: LIGASE Ligands: ATP, ALA, CIT, TRS, GOL |
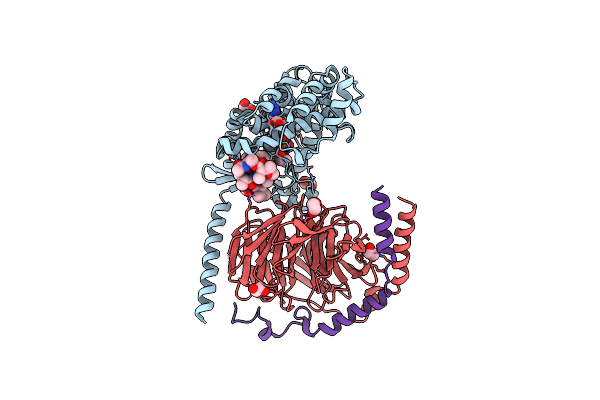 |
Crystal Structure Of The G11 Protein Heterotrimer Bound To Ym-254890 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: GDP, EDO, DAM, HF2, THC, OTH, HL2, MAA, ALA, ACE |
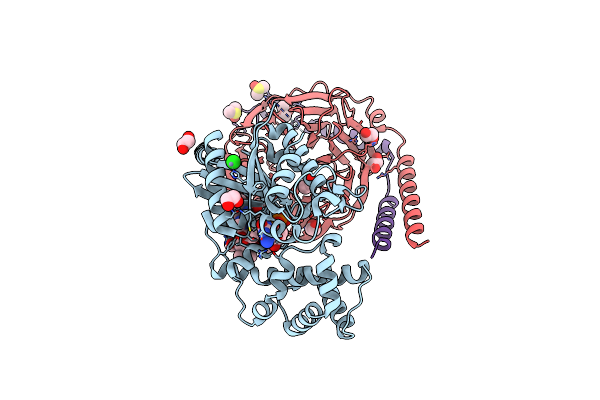 |
Crystal Structure Of The G11 Protein Heterotrimer Bound To Fr900359 Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2025-03-19 Classification: SIGNALING PROTEIN Ligands: EDO, GDP, ZN, CL, DAM, HF2, UDL, OTH, HL2, MAA, ALA, PPI, DMS |
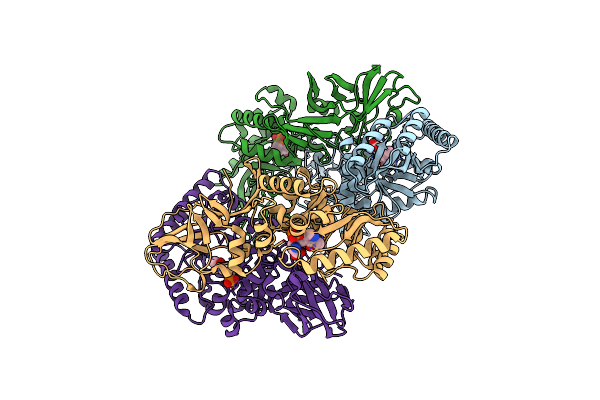 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
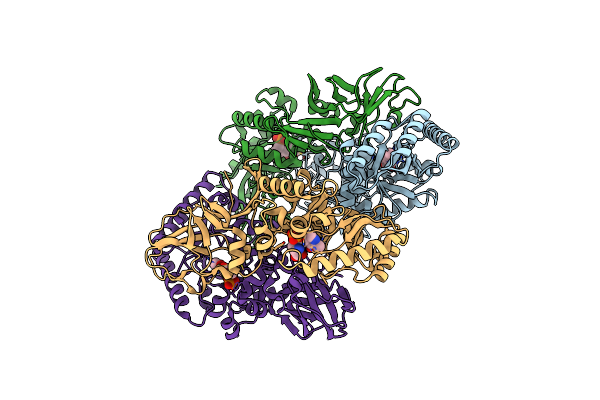 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, ALA, CL |
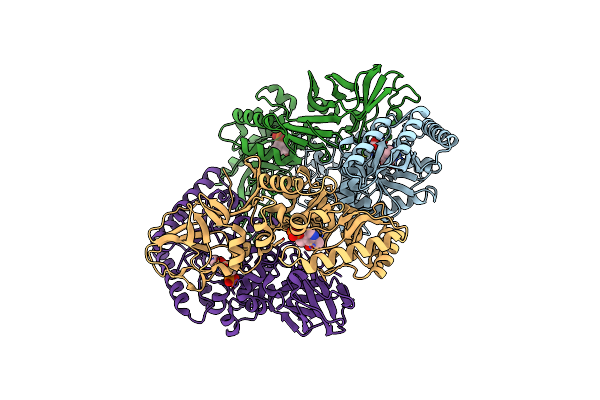 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
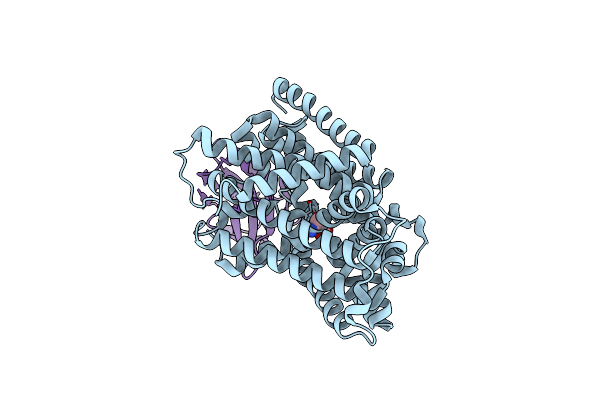 |
Organism: Carnobacterium sp. at7, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: MEMBRANE PROTEIN Ligands: ALA |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-01 Classification: ISOMERASE Ligands: PLP, CL, ALA |
 |
Structure Of Wild-Type Aminotransferase From Mycolicibacterium Neoaurum In Complex With Llp And Ala
Organism: Mycolicibacterium neoaurum vkm ac-1815d
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2024-12-25 Classification: TRANSFERASE Ligands: ALA |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-12-11 Classification: PROTEIN BINDING Ligands: PHE, ALA, NI, MG |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-11 Classification: FLAVOPROTEIN Ligands: ALA, FAD, GLY |
 |
Organism: Methanothermobacter phage psim100
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-10-09 Classification: HYDROLASE Ligands: DGL, ALA |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Cchacs From Conidiobolus Coronatus In The Complex With Thdp And Adp
Organism: Conidiobolus coronatus nrrl 28638
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-10-02 Classification: LYASE Ligands: TPP, ADP, EDO, ALA, ACT, MG, CL, GLY, FMT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: MEMBRANE PROTEIN Ligands: NAG, ALA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: PROTEIN TRANSPORT Ligands: ALA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: PROTEIN TRANSPORT Ligands: ALA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: PROTEIN TRANSPORT Ligands: ALA |
 |
Escherichia Coli Paused Disome Complex (Non-Rotated Disome Interface Class 1)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-03-13 Classification: RIBOSOME Ligands: ZN, PUT, SPD, MG, ALA |

