Search Count: 25
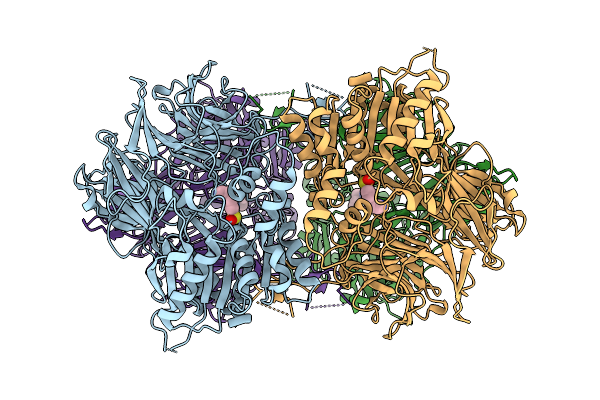 |
Cryo-Em Structure Of Acylaminoacyl Peptidase (Aap) In Covalent Complex With Inhibitor Aebsf
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE Ligands: AES |
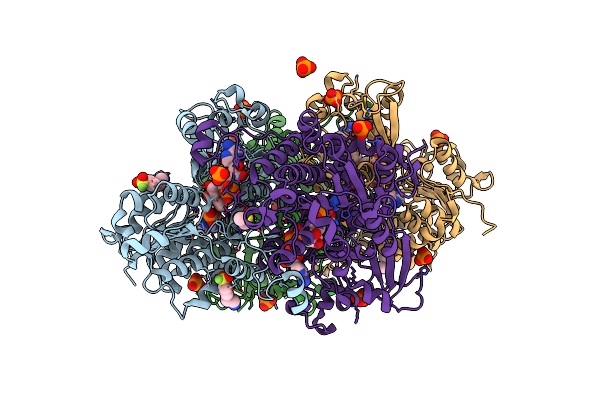 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From P. Aeruginosa In Complex With F2X-Entry Library Fragment F11
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: NAD, ADE, K, PO4, AES, DMS |
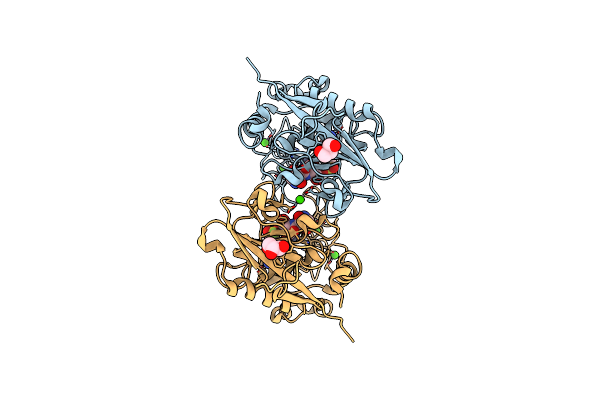 |
Organism: Methanosarcina acetivorans c2a, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-03-22 Classification: HYDROLASE Ligands: CA, ZN, AES, GOL |
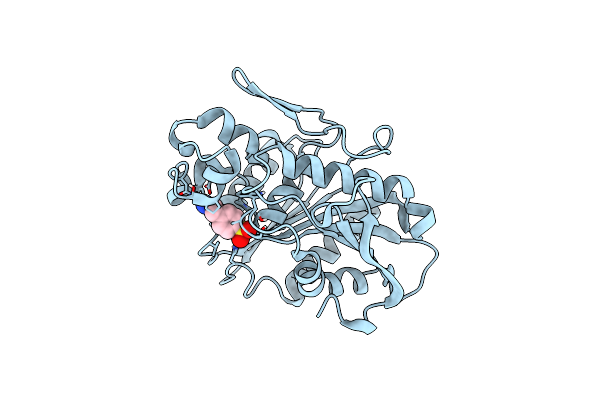 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: AES |
 |
Pandda Analysis Group Deposition -- Endothiapepsin Changed State Model For Fragment F2X-Entry Library F11A
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2020-06-03 Classification: HYDROLASE Ligands: AES, DMS, GOL, ACT, PG4 |
 |
Organism: Lysobacter sp. (strain xl1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: SO4, AES, GOL, PGE, JAT, CL, PEG |
 |
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-08-08 Classification: LYASE Ligands: AES, SO4, CA |
 |
Organism: Lysobacter sp. (strain xl1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-01-17 Classification: HYDROLASE Ligands: SO4, AES, GOL, PGE, CL, PEG |
 |
Organism: Chaetomium thermophilum var. thermophilum dsm 1495, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-03-02 Classification: HYDROLASE Ligands: MG, AGS, AES |
 |
X-Ray Structure Of Psaa From Yersinia Pestis, In Complex With Galactose And Aebsf
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-05-22 Classification: CELL ADHESION/inhibitor Ligands: GAL, BR, GAI, AES, GLY, MLI |
 |
X-Ray Structure Of Psaa From Yersinia Pestis, In Complex With Lactose And Aebsf
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-05-22 Classification: CELL ADHESION/inhibitor Ligands: GAI, CL, AES |
 |
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-01-02 Classification: MEMBRANE PROTEIN Ligands: AES |
 |
Structural Investigation Of Bacteriophage Phi6 Lysin (In Complex With Chitotetraose)
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2013-01-02 Classification: HYDROLASE Ligands: AES |
 |
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-09-05 Classification: HYDROLASE Ligands: GOL, CL, 1PE, AES, PEG, PGE |
 |
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-09-05 Classification: HYDROLASE Ligands: GOL, P33, CL, AES, PG4, PG0 |
 |
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-09-05 Classification: HYDROLASE Ligands: GOL, PG0, PG4, AES, 1PE, CL |
 |
Crystal Structure Of The N-Terminal Beta-Aminopeptidase Bapa In Complex With Pefabloc Sc (Aebsf)
Organism: Sphingosinicella xenopeptidilytica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-06-08 Classification: Hydrolase/Hydrolase Inhibitor Ligands: GOL, EPE, AES, SO4 |
 |
A Second Transient Position Of Atp On Its Trail To The Nucleotide-Binding Site Of Subunit B Of The Motor Protein A1Ao Atp Synthase
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:3.43 Å Release Date: 2009-02-10 Classification: HYDROLASE Ligands: ATP, AES |
 |
Intermediate Position Of Atp On Its Trail To The Binding Pocket Inside The Subunit B Mutant R416W Of The Energy Converter A1Ao Atp Synthase
Organism: Methanosarcina mazei
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-09-09 Classification: HYDROLASE Ligands: ATP, AES, CIT |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2007-07-03 Classification: hydrolase/hydrolase inhibitor Ligands: SO4, AES, GOL |

