Search Count: 29
 |
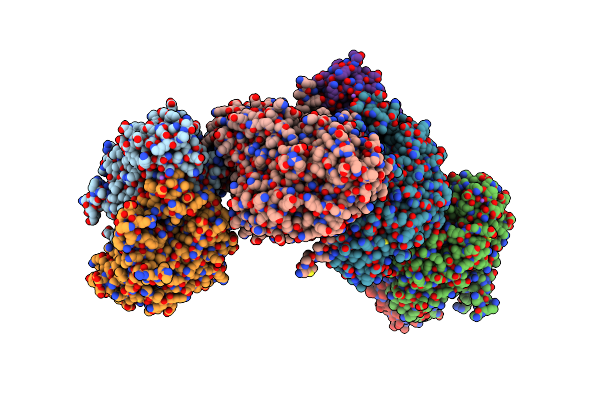
Crystal Structure Of A Polyketide Cyclase Fasu From Streptomyces Kanamyceticus
Organism: Streptomyces kanamyceticus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-03-12 Classification: BIOSYNTHETIC PROTEIN Ligands: AE3, MG |
 |
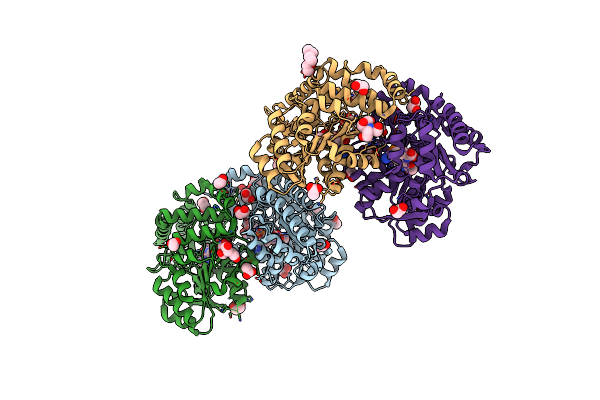
Promiscuous Amino Acid Gamma Synthase From Caldicellulosiruptor Hydrothermalis In Closed Conformation
Organism: Caldicellulosiruptor hydrothermalis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: MES, NA, AE3, ETE, 1PE |
 |
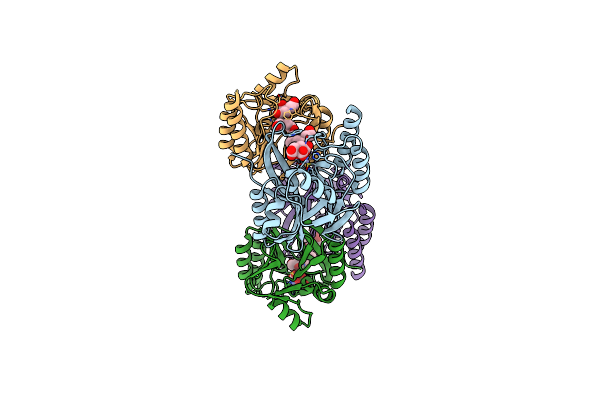
Organism: Pseudoalteromonas luteoviolacea
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-10-06 Classification: BIOSYNTHETIC PROTEIN Ligands: BTB, ACT, EJ1, GOL, EDO, AE3 |
 |
Crystal Structure Of Metallo-Beta-Lactamase Imp-1 Mutant (D120E) In Complex With Citrate.
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-08-04 Classification: HYDROLASE Ligands: ZN, FLC, AE3 |
 |
Crystal Structure Of Gh5_18 From Streptomyces Cattleya In Complex With Glcnac
Organism: Streptomyces cattleya (strain atcc 35852 / dsm 46488 / jcm 4925 / nbrc 14057 / nrrl 8057)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-04-07 Classification: HYDROLASE Ligands: NAG, AE3, EDO |
 |
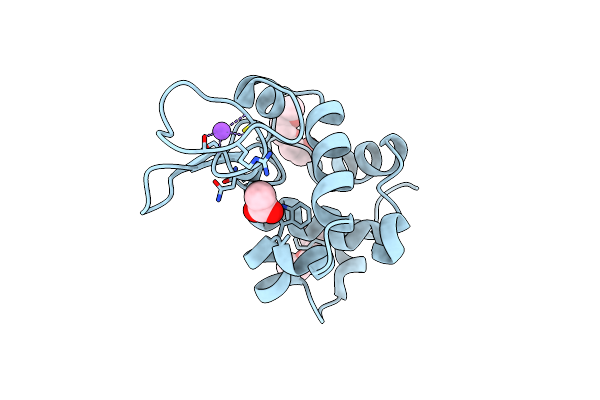
Crystal Structure Of Gh5_18-E153A From Streptomyces Cattleya In Complex With Manb1-4Glcnac
Organism: Streptomyces cattleya (strain atcc 35852 / dsm 46488 / jcm 4925 / nbrc 14057 / nrrl 8057)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-04-07 Classification: HYDROLASE Ligands: AE3, EDO, SO4 |
 |
Structure Of Lysozyme From Sin Imisx Setup Collected By Rotation Serial Crystallography On Crystals Prelocated By 2D X-Ray Phase-Contrast Imaging
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: BR, NA, ACY, PG0, AE3 |
 |
Structure Of Lysozyme From Sin Imisx Setup Collected By Still Serial Crystallography On Crystals Prelocated By 2D X-Ray Phase-Contrast Imaging
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: AE3, BR, NA, ACY |
 |
Crystal Structure Of A Peptidoglycan Release Complex, Sagb-Spdc, In Lipidic Cubic Phase
Organism: Staphylococcus aureus (strain bovine rf122 / et3-1), Staphylococcus aureus (strain nctc 8325), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-09-23 Classification: HYDROLASE Ligands: FLC, PG4, OLC, SO4, AE3 |
 |
Crystal Structure Of Thermotoga Maritima Threonylcarbamoyladenosine Biosynthesis Complex Tsab2D2E2 Bound To Atp And Carboxy-Amp
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-05-22 Classification: BIOSYNTHETIC PROTEIN Ligands: PGE, AE3, KG4, ZN, ADP, ATP, MG |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2019-03-27 Classification: LIPID TRANSPORT Ligands: CL, CA, AE3, PG4, PEG, GOL, LMT, JU7, MA4 |
 |
Organism: Dictyoglomus thermophilum (strain atcc 35947 / dsm 3960 / h-6-12)
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: PGE, AE3 |
 |
Crystal Structure Of The Tandem Duf26 Ectodomain From The Arabidopsis Thaliana Cysteine-Rich Receptor-Like Protein Pdlp5.
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2018-12-26 Classification: SIGNALING PROTEIN Ligands: NAG, EDO, NA, SO4, GOL, AE3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-12 Classification: IMMUNE SYSTEM Ligands: CL, AE3, PG4 |
 |
Plasmodium Falciparum 6-Phosphogluconate Dehydrogenase In Its Apo Form, In Complex With Its Cofactor Nadp+ And In Complex With Its Substrate 6-Phosphogluconate
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-08-22 Classification: OXIDOREDUCTASE Ligands: GOL, PEG, PGE, AE3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-01-10 Classification: IMMUNE SYSTEM Ligands: PG0, EDO, PEG, AE3 |
 |
Structure Of 4-Hydroxytetrahydrodipicolinate Reductase From Mycobacterium Tuberculosis With Nadph And 2,6 Pyridine Dicarboxylic Acid
Organism: Mycobacterium tuberculosis (strain atcc 25177 / h37ra)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-01-10 Classification: OXIDOREDUCTASE Ligands: PDC, BEZ, IMD, NAP, AE3, CL, EDO, SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-09-06 Classification: HYDROLASE Ligands: NAG, PG0, ETX, EDO, CVI, PEG, PGE, AE3, PE4 |
 |
Structure Of A Pre-Reaction Ternary Complex Between Sarin- Acetylcholinesterase And Hi-6
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-05-11 Classification: HYDROLASE Ligands: NAG, CO3, P6G, AE3, HI6 |
 |
Crystal Structure Of Betaine Aldehyde Dehydrogenase From Spinach Showing A Thiohemiacetal With Betaine Aldehyde
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2016-02-03 Classification: OXIDOREDUCTASE Ligands: CHT, ETX, K, PG4, AE3, GOL |

