Search Count: 6
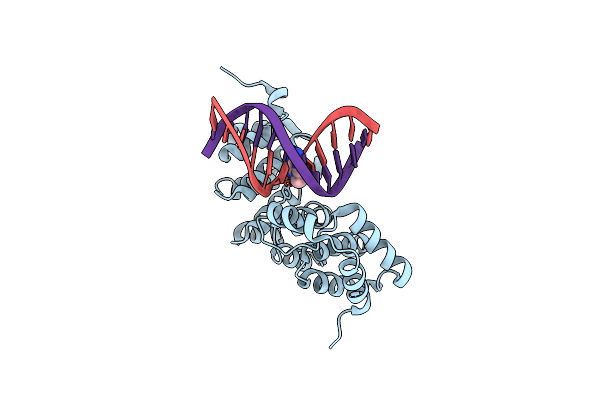 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing An Oxocarbenium-Intermediate Analog And A Free 3-Methyladenine Nucleobase
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: ADK |
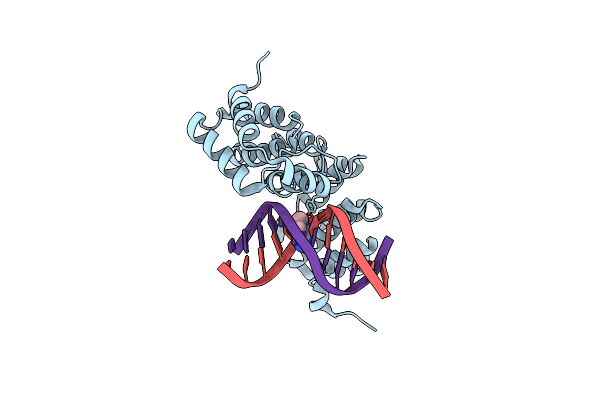 |
Alkylpurine Dna Glycosylase Alkd Bound To Dna Containing An Abasic-Site Analog And A Free 3-Methyladenine Nucleobase
Organism: Bacillus cereus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2015-10-28 Classification: HYDROLASE/DNA Ligands: ADK |
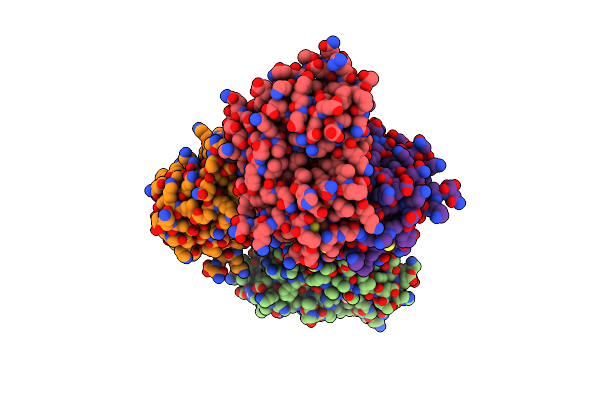 |
The Structural Basis Of 3-Methyladenine Recognition By 3- Methyladenine Dna Glycosylase I (Tag) From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-02-22 Classification: HYDROLASE Ligands: ZN, ADK, SO4 |
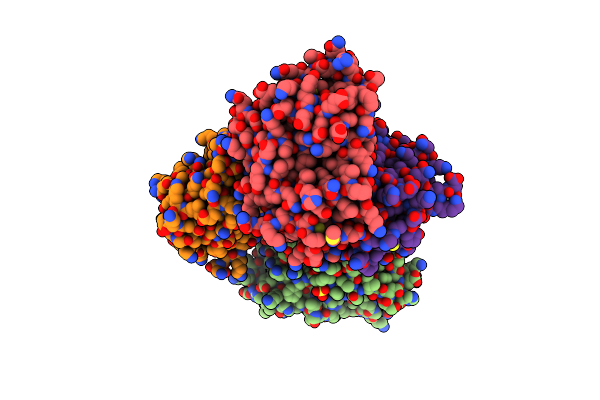 |
Crystal Structure Of Y16F Of 3-Methyladenine Dna Glycosylase I (Tag) In Complex With 3-Methyladenine
Organism: Staphylococcus aureus subsp. aureus mssa476
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2012-02-15 Classification: HYDROLASE Ligands: ZN, ADK, SO4 |
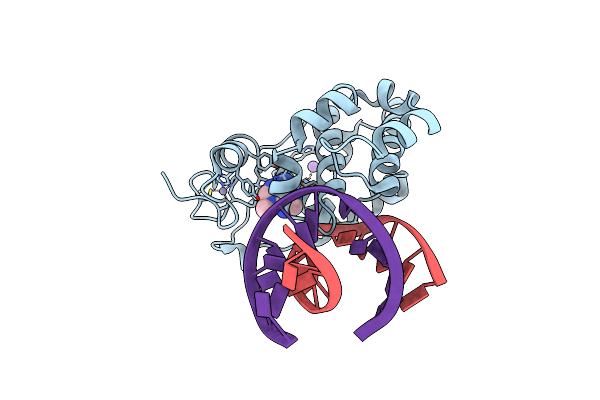 |
Crystal Structure Of 3-Methyladenine Dna Glycosylase I (Tag) Bound To Dna/3Ma
Organism: Salmonella typhi
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-05-15 Classification: 3-METHYLADENINE DNA GLYCOSYLASE I/DNA Ligands: ZN, NA, ADK |
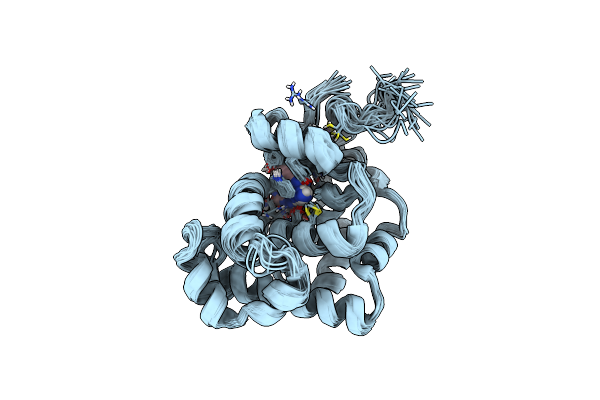 |
Solution Structure And Base Perturbation Studies Reveal A Novel Mode Of Alkylated Base Recognition By 3-Methyladenine Dna Glycosylase I
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2003-11-25 Classification: HYDROLASE Ligands: ZN, ADK |

