Search Count: 200
All
Selected
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix 100K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2026-03-04 Classification: TRANSFERASE Ligands: PO4, ADE, PGE, PEG |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix 323K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2026-02-18 Classification: TRANSFERASE Ligands: PO4, ADE, PEG |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix 293K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2026-02-18 Classification: TRANSFERASE Ligands: PO4, PEG, ADE |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix 343K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-02-11 Classification: TRANSFERASE Ligands: PO4, PEG, ADE |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix 333K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2026-02-11 Classification: TRANSFERASE Ligands: PO4, PEG, ADE |
 |
Crystal Structure Of 5'-Deoxy-5'-Methylthioadenosine Phosphorylase From Aeropyrum Pernix 353K
Organism: Aeropyrum pernix k1
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: PO4, ADE |
 |
Structure Of Kluyveromyces Lactis Mrna Cap (Guanine-N7) Methyltransferase, Abd1, In Complex With Adenine And M7Gtp
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: ADE, MGP, P4G, SO4, EDO |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2025-04-16 Classification: DNA Ligands: ADE, NA |
 |
Structure Of The Wild-Type Azg2 In Arabidopsis Thaliana In The Adenine-Bound State At Ph 5.5
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN Ligands: ADE |
 |
Structure Of The Wild-Type Azg2 In Arabidopsis Thaliana In The Adenine-Bound State At Ph 7.4
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: TRANSPORT PROTEIN Ligands: ADE |
 |
Organism: Streptantibioticus cattleyicolor
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2025-03-05 Classification: LIGASE Ligands: ADE |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Trichomonas Vaginalis (Phosphate/Adenine Bound)
Organism: Trichomonas vaginalis g3
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: CL, PO4, ADE |
 |
Organism: Kitasatospora mediocidica kctc 9733
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: ADE, HEM, UYM |
 |
Organism: Kitasatospora mediocidica kctc 9733
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-11-20 Classification: OXIDOREDUCTASE Ligands: HEM, ADE |
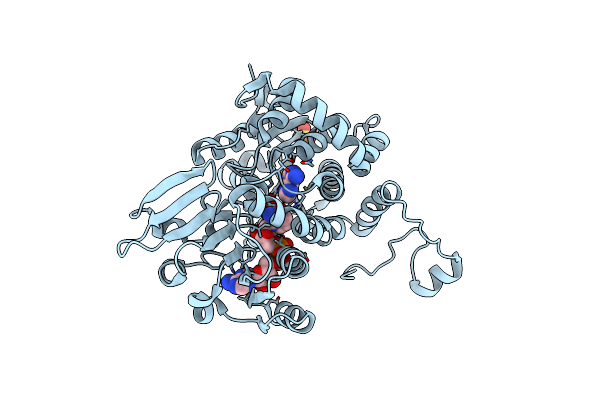 |
The Crystal Structure Of Legionella Pneumophila Adenosylhomocysteinase Lpg2021 In Ternary Complex With Nad And Adenine
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: NAD, ADE, SO4 |
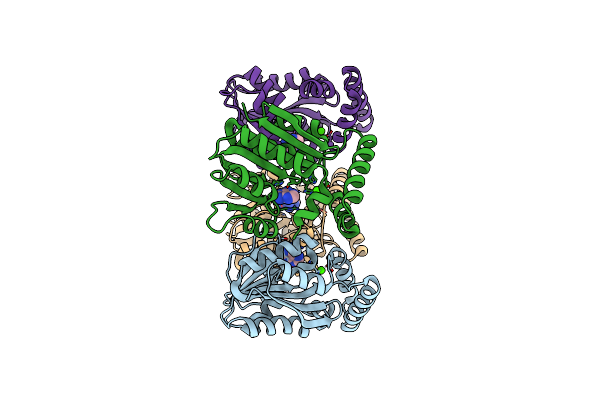 |
Organism: Kitasatospora cystarginea
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-08-21 Classification: TRANSFERASE Ligands: ADE, CA |
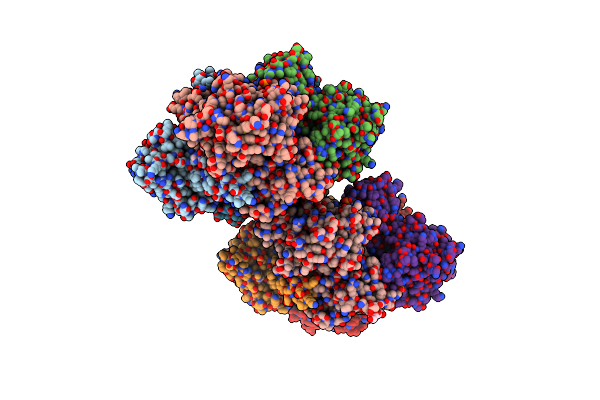 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From P. Aeruginosa In Complex With F2X-Entry Library Fragment H09
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-05-22 Classification: HYDROLASE Ligands: NAD, ADE, PO4, K, A1H8H, GOL |
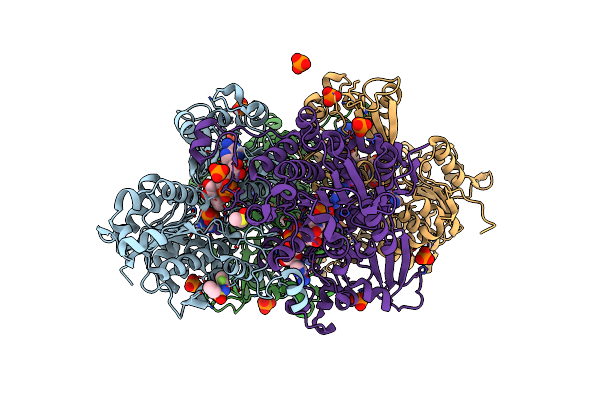 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From P. Aeruginosa In Complex With F2X-Entry Library Fragment D02
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: NAD, ADE, DMS, K, PO4, SY4 |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From P. Aeruginosa In Complex With F2X-Entry Library Fragment A01
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: NAD, ADE, K, PO4, UHR, DMS |
 |
Crystal Structure Of S-Adenosyl-L-Homocysteine Hydrolase From P. Aeruginosa In Complex With F2X-Entry Library Fragment A07
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-02-21 Classification: HYDROLASE Ligands: NAD, ADE, K, PO4, DMS, R8A |

