Search Count: 42
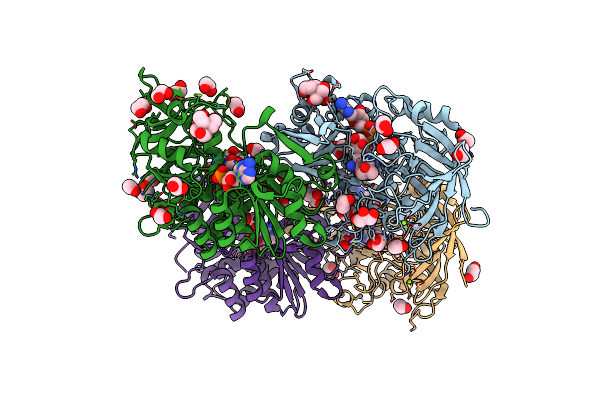 |
Crystal Structure Of Hydroxynitrile Lyase From Linum Usitatissimum Complexed With Acetone Cyanohydrin
Organism: Linum usitatissimum
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-02-09 Classification: LYASE Ligands: NAD, ZN, CNH, MG, EDO, GOL, BTB, ACN |
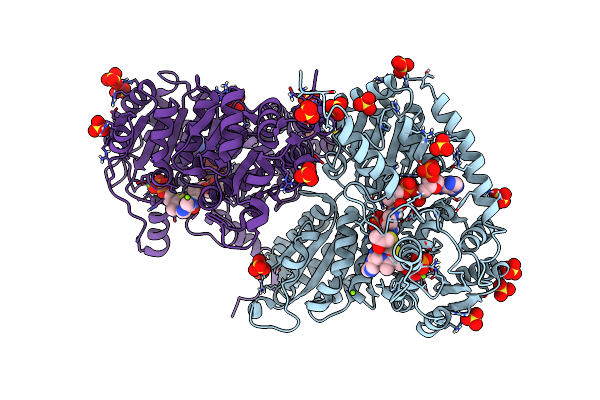 |
Actinobacterial 2-Hydroxyacyl-Coa Lyase (Achacl) Structure In Complex With A Covalently Bound Reaction Intermediate As Well As Products Formyl-Coa And Acetone
Organism: Actinomycetospora chiangmaiensis dsm 45062
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-02-02 Classification: LYASE Ligands: TPP, SO4, ACN, FYN, MG, OXT |
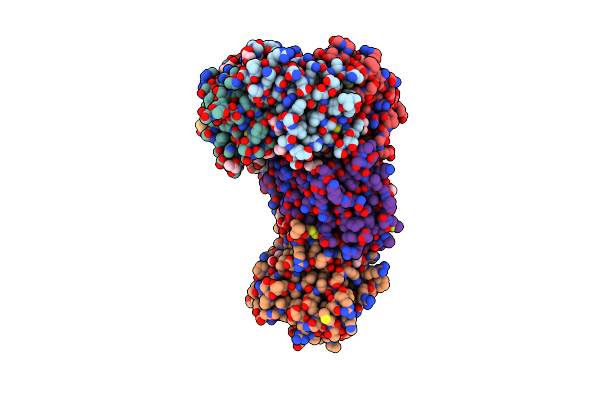 |
Organism: Salmonella enterica subsp. enterica serovar typhi str. ct18
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2021-12-22 Classification: TOXIN Ligands: PEG, ACN, GOL, FLC |
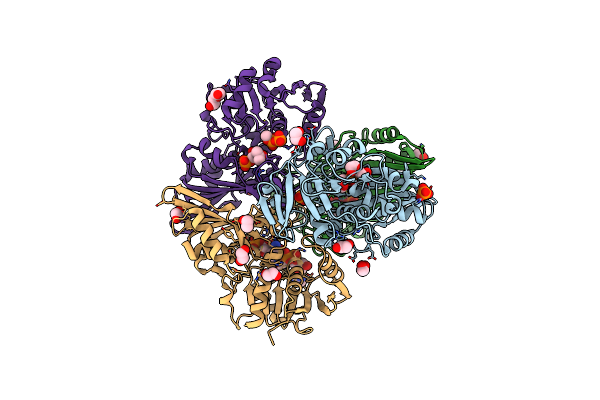 |
Crystal Structure Of Thioacyl-Glyceraldehyde-3-Phosphate Dehydrogenase 1(Gapdh 1) From Escherichia Coli At 1.77 Angstrom Resolution
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-05-26 Classification: OXIDOREDUCTASE Ligands: G3H, PO4, EDO, PEG, ACN, NAD |
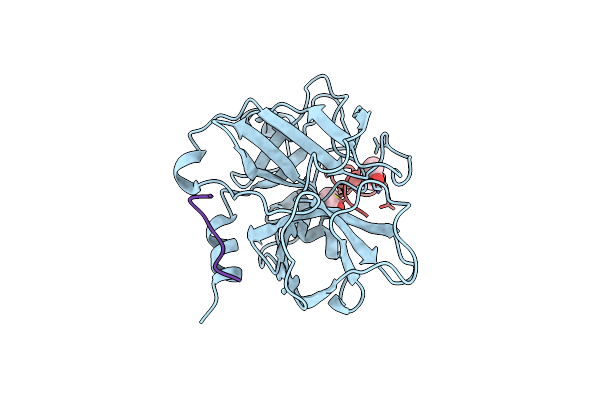 |
Crystal Structure Of The Catalytic Domain Of The Coagulation Factor Xia In Complex With Double Bridged Peptide F21
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2020-05-20 Classification: BLOOD CLOTTING Ligands: ACN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-11-13 Classification: HORMONE Ligands: IPH, ACN, ZN, CL, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2019-02-20 Classification: Transferase/Transferase Inhibitor Ligands: JRD, MES, ACN |
 |
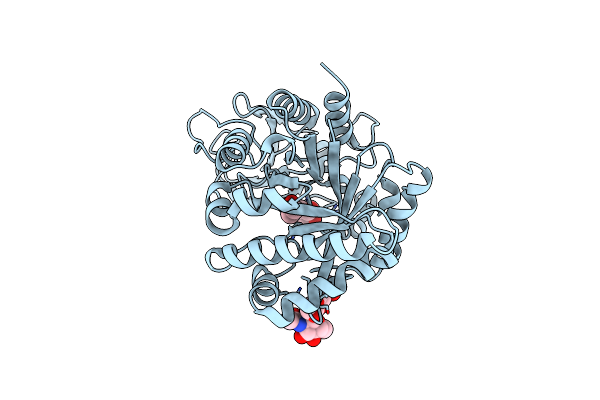
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2018-02-21 Classification: HORMONE Ligands: IPH, ZN, CL, NA, GOL, ACN |
 |
Crystal Structure Of Signalling Protein From Buffalo (Spb-40) With An Acetone Induced Conformation Of Trp78 At 1.49 A Resolution
Organism: Bubalus bubalis
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2018-01-31 Classification: SIGNALING PROTEIN Ligands: MPD, ACN |
 |
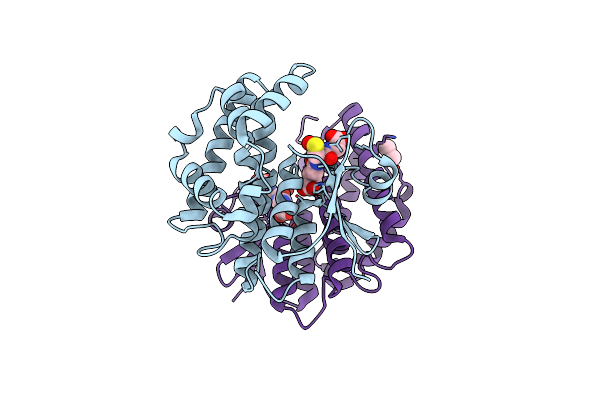
Crystal Structure Of 7,8-Diaminopelargonic Acid Synthase (Bioa) From Mycobacterium Tuberculosis Complexed With A Dsf Fragment Hit
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2015-07-08 Classification: transferase/transferase inhibitor Ligands: PLP, 3VW, EPE, SO4, ACN |
 |
Crystal Structure Of A Tau Class Glutathione Transferase 10 From Glycine Max
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-12-17 Classification: TRANSFERASE Ligands: GS8, ACN |
 |
Crystal Structure Of An Immunoreactive 32 Kda Antigen Pg49 (Pg_0181) From Porphyromonas Gingivalis W83 At 1.45 A Resolution (Psi Community Target, Nakayama)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-12-03 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: EDO, ACN |
 |
Crystallographic Structure Analysis Of Urease From Jack Bean (Canavalia Ensiformis) At 1.49 A Resolution
Organism: Canavalia ensiformis
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2012-11-07 Classification: HYDROLASE Ligands: NI, PO4, EDO, ACN, BME, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-08-08 Classification: TRANSFERASE Ligands: ZN, TCE, TAM, ACN |
 |
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp. r39
Method: X-RAY DIFFRACTION Release Date: 2012-02-29 Classification: HYDROLASE Ligands: SO4, MG, FP5, ACN, 33D, MES |
 |
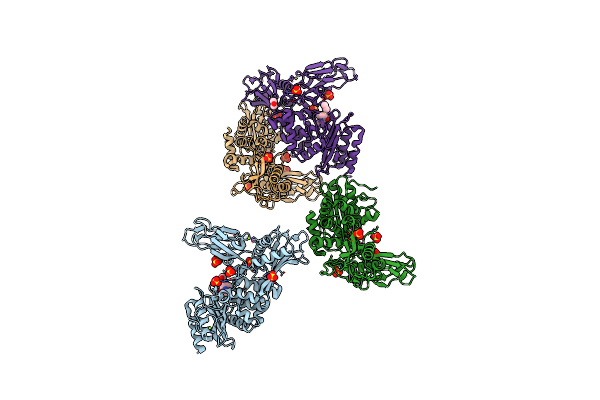
Crystal Structure Of Anthrax Protective Antigen Mutant S337C N664C And Dithiolacetone Modified To 1.8-A Resolution
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2011-10-26 Classification: PROTEIN TRANSPORT, TOXIN Ligands: CA, MXE, ACN |
 |
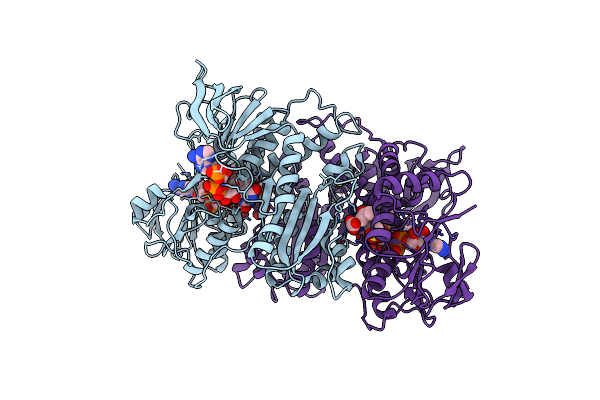
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp. r39
Method: X-RAY DIFFRACTION Release Date: 2011-07-27 Classification: HYDROLASE Ligands: FP5, SO4, MG, ACN |
 |
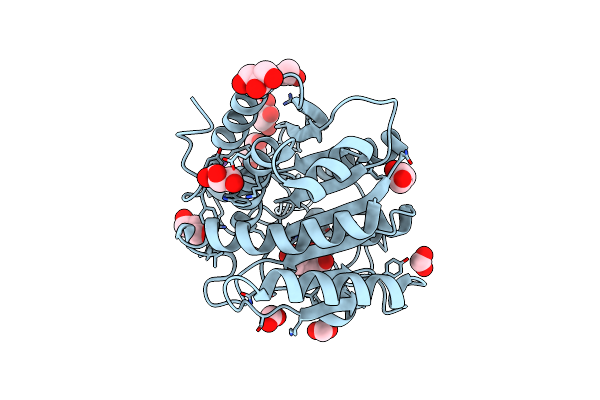
Structural Basis For Carbon Dioxide Binding By 2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2011-02-16 Classification: OXIDOREDUCTASE Ligands: NAP, FAD, KPC, MG, BCT, CO2, ACN, COM |
 |
Carboxypeptidase A Liganded To An Organic Small-Molecule: Conformational Changes
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-10-27 Classification: HYDROLASE Ligands: ZN, NA, CIT, ACN, FMT, GOL |
 |
Crystal Structure Of The First Plant Urease From Jack Bean (Canavalia Ensiformis)
Organism: Canavalia ensiformis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-07-28 Classification: HYDROLASE Ligands: NI, PO4, ACN |

