Search Count: 28
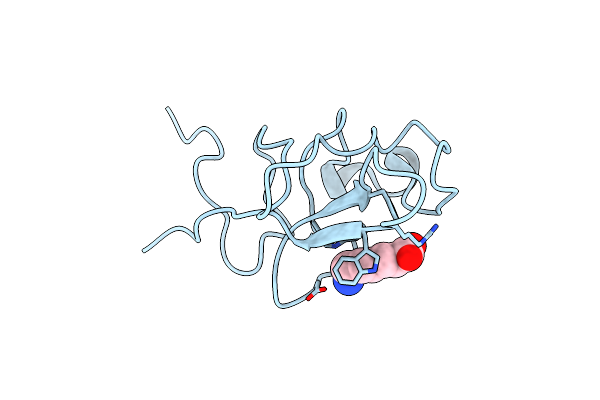 |
The Atomic Resolution Crystal Structure Of Kringle 2 Variant Bound With Eaca
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2019-05-15 Classification: BLOOD CLOTTING Ligands: ACA |
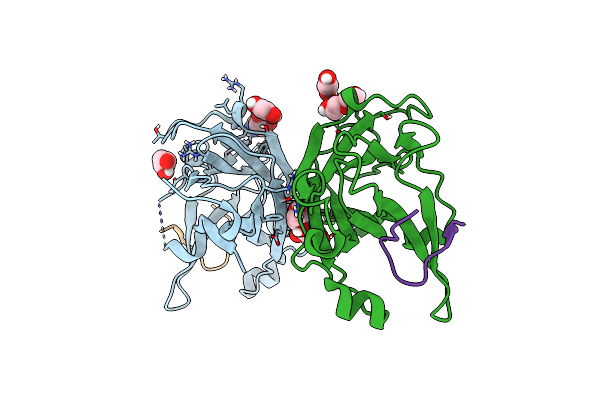 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2019-05-01 Classification: TRANSFERASE Ligands: GOL, ACA, EDO, PEG |
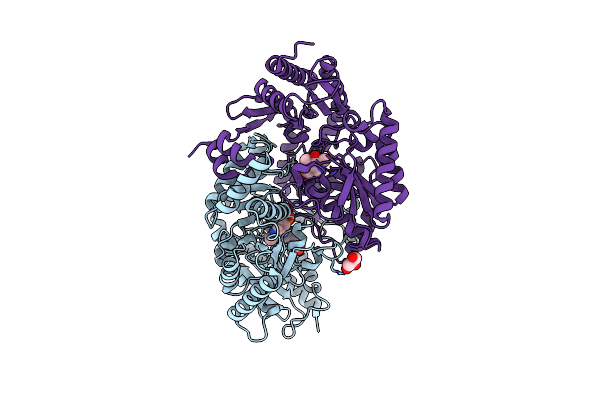 |
Crystal Structure Of The Omega Transaminase From Pseudomonas Jessenii In Complex With Plp And 6-Aminohexanoate (6-Aca)
Organism: Pseudomonas sp
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2019-04-10 Classification: TRANSFERASE Ligands: PLP, ACA, GOL |
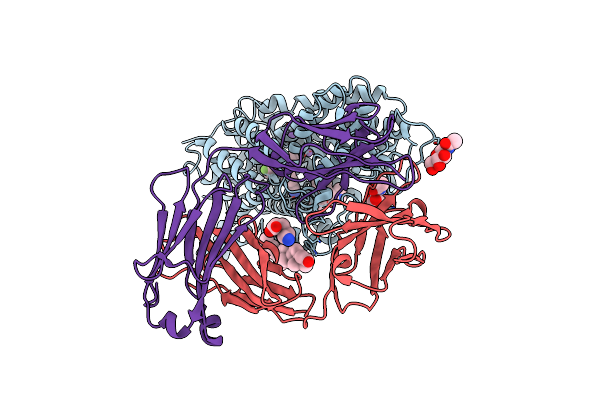 |
X-Ray Structure Of The Ts3 Human Serotonin Transporter Complexed With S-Citalopram At The Central And Allosteric Sites
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2016-04-13 Classification: MEMBRANE PROTEIN Ligands: 68P, NAG, CLR, C14, NA, ACA |
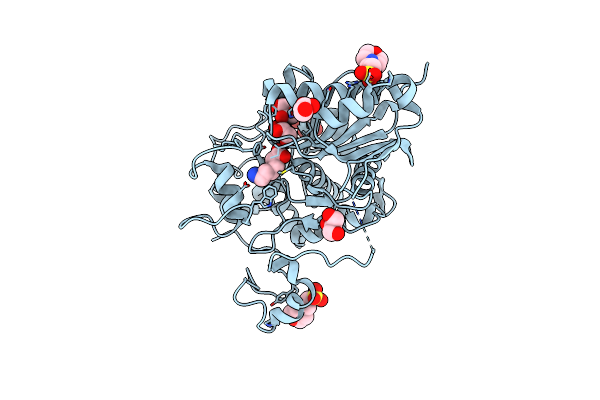 |
6-Aminohexanoate-Dimer Hydrolase S112A/G181D/R187A/H266N/D370Y Mutant Complexd With 6-Aminohexanoate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: ACA, GOL, MES |
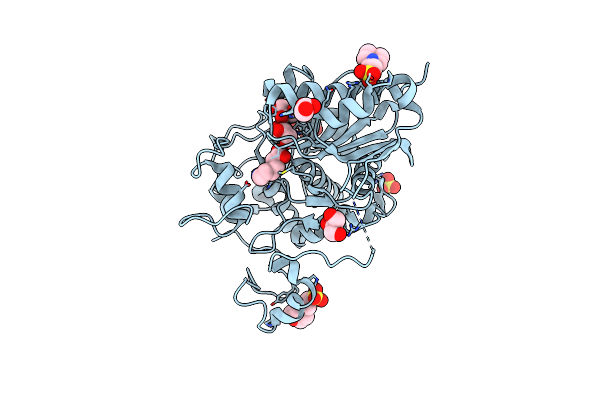 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/R187G/H266N/D370Y Mutant Complexd With 6-Aminohexanoate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-10-16 Classification: HYDROLASE Ligands: ACA, GOL, MES, SO4 |
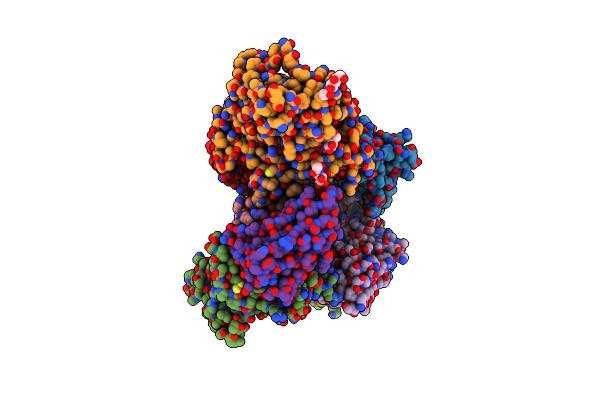 |
Crystal Structure Of Hiv-1 Yu2 Envelope Gp120 Glycoprotein In Complex With Cd4-Mimetic Miniprotein, M48U1, And Llama Single-Domain, Broadly Neutralizing, Co-Receptor Binding Site Antibody, Jm4
Organism: Human immunodeficiency virus 1, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2013-08-14 Classification: VIRAL PROTEIN/INHIBITOR Ligands: NAG, EDO, ACA |
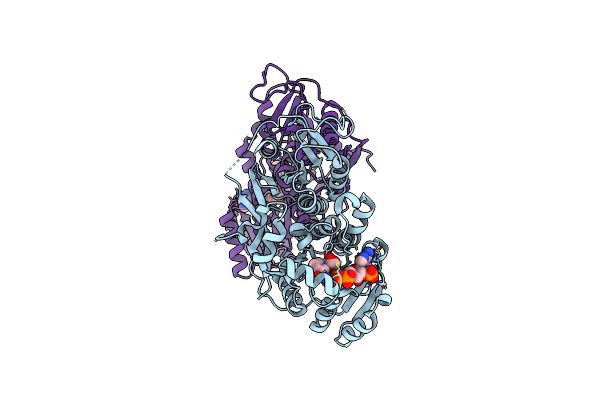 |
Plmkr1-Ketoreductase From The First Module Of Phoslactomycin Biosynthesis In Streptomyces Sp. Hk803
Organism: Streptomyces sp. hk803
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2013-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, ACA |
 |
Crystal Structure Of Proteinaceous Rnase P 1 (Prorp1) From A. Thaliana With Mn
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-09-26 Classification: RNA BINDING PROTEIN Ligands: ZN, MN, ACA |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-07-18 Classification: TRANSCRIPTION Ligands: ACA |
 |
Crystal Structure Of C-Lobe Of Bovine Lactoferrin Complexed With Aminocaproic Acid At 1.46 A Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2012-03-14 Classification: METAL BINDING PROTEIN Ligands: NAG, FE, ZN, CO3, SO4, ACA |
 |
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/H266N Mutant With Substrate
Organism: Flavobacterium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-09-01 Classification: HYDROLASE Ligands: ACA, MES, GOL, NA |
 |
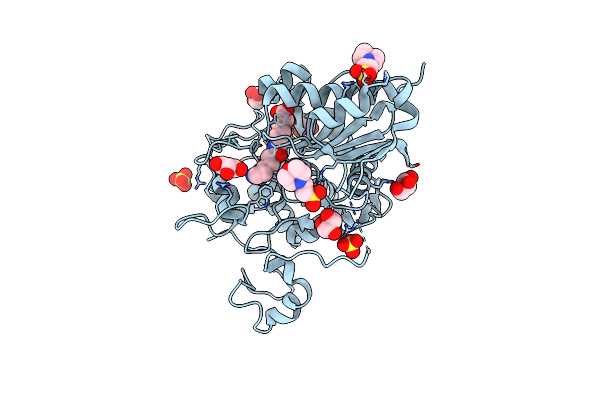
Structure Of 6-Aminohexanoate Cyclic Dimer Hydrolase Complexed With Substrate
Organism: Arthrobacter sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-11-03 Classification: HYDROLASE Ligands: ACA, GOL |
 |
Structure Of 6-Aminohexanoate-Dimer Hydrolase, S112A/D370Y Mutant Complexed With 6-Aminohexanoate-Dimer
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-04-14 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
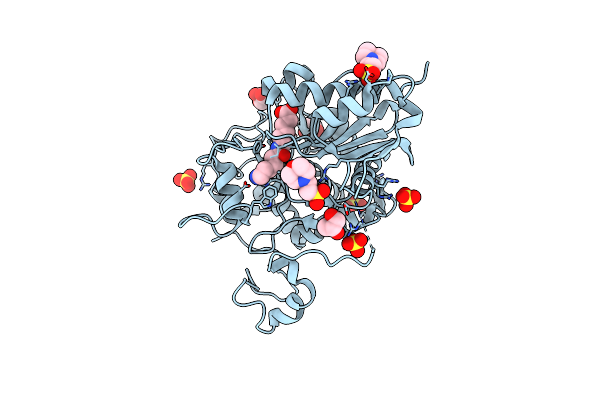
Structure Of 6-Aminohexanoate-Dimer Hydrolase, A61V/S112A/A124V/R187S/F264C/G291R/G338A/D370Y Mutant (Hyb-S4M94) With Substrate
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-04-14 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
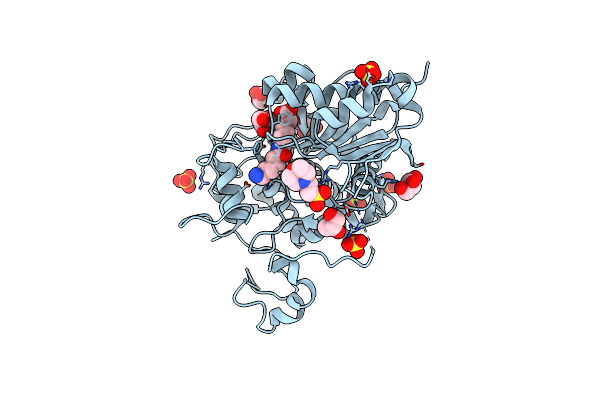
Structure Of 6-Aminohexanoate-Dimer Hydrolase, S112A/G181D Mutant Complexed With 6-Aminohexanoate-Dimer
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-04-07 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
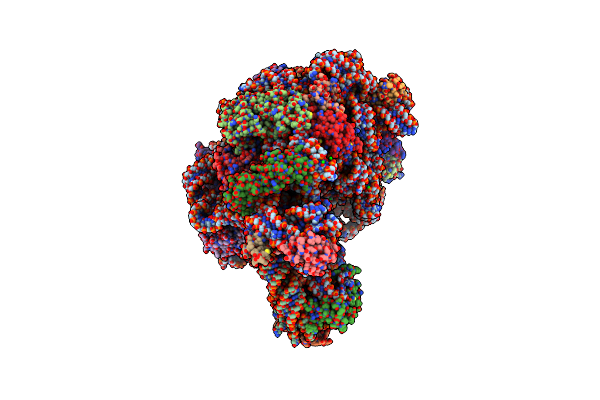
Crystal Structure Of 6-Aminohexanoate-Dimer Hydrolase S112A/G181D/H266N/D370Y Mutant With Substrate
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2009-04-07 Classification: HYDROLASE Ligands: SO4, ACA, MES, GOL |
 |
The Structure Of Cca And Cca-Phe-Cap-Bio Bound To The Large Ribosomal Subunit Of Haloarcula Marismortui
Organism: Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-09-23 Classification: RIBOSOME Ligands: MG, CL, SR, NA, CD, K, PHE, ACA |
 |
The Structure Of Ca And Cca-Phe-Cap-Bio Bound To The Large Ribosomal Subunit Of Haloarcula Marismortui
Organism: Synthetic construct, Haloarcula marismortui
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2008-09-23 Classification: RIBOSOME Ligands: MG, CL, SR, NA, CD, K, PHE, ACA |
 |
Nmr Structure Of Immunosuppressory Peptide Containing Cyclolinopeptide X And Antennapedia(43-58) Sequences
|

