Search Count: 21
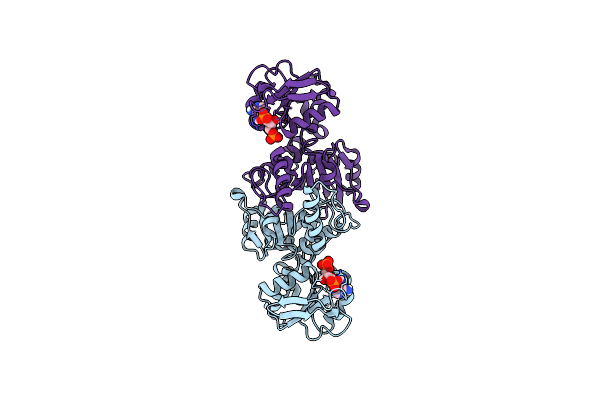 |
Crystal Structure Of Aspartate Semialdehyde Dehydrogenase From Porphyromonas Gingivalis Complexed With 2',5'Adenosine Diphosphate
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2024-06-05 Classification: OXIDOREDUCTASE Ligands: A2P |
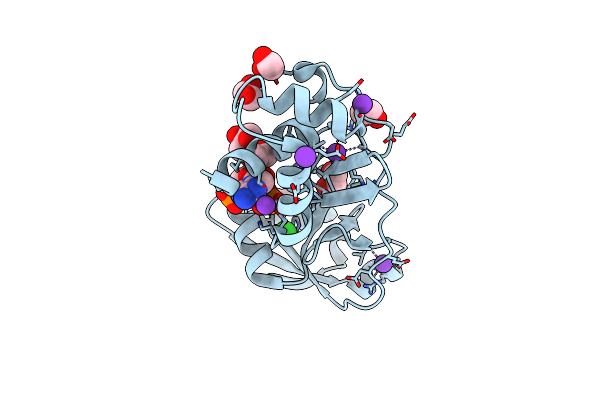 |
Trna 2'-Phosphotransferase (Tpt1) From Pyrococcus Horikoshii In Complex With 2',5'-Adp
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-08 Classification: TRANSFERASE Ligands: A2P, EDO, GOL, CL, K |
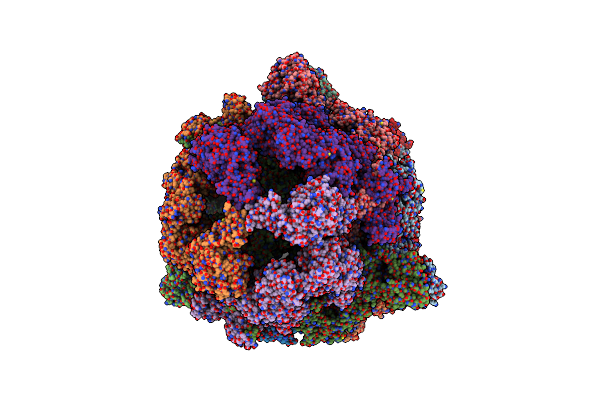 |
Structure Of Fatty Acid Synthase Complex From Saccharomyces Cerevisiae At 2.9 Angstrom
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2020-03-18 Classification: TRANSFERASE Ligands: PNS, EDO, NA, A2P, ACT, PGE, FMN, MLI |
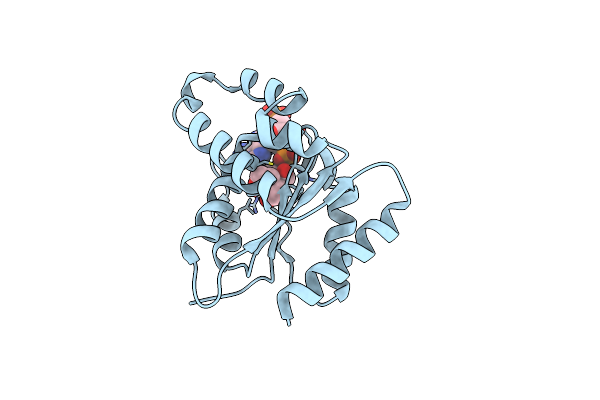 |
Crystal Structure Of The Tir Domain From The Grapevine Disease Resistance Protein Run1 In Complex With Nadp+ And Bis-Tris
Organism: Vitis rotundifolia
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-09-04 Classification: SIGNALING PROTEIN Ligands: A2P, BTB |
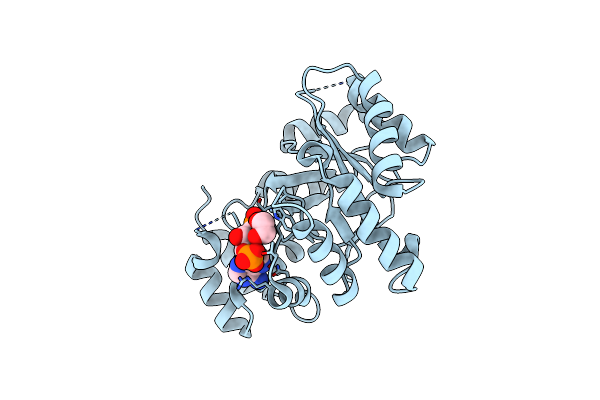 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2016-11-09 Classification: OXIDOREDUCTASE Ligands: A2P, EDO, CL |
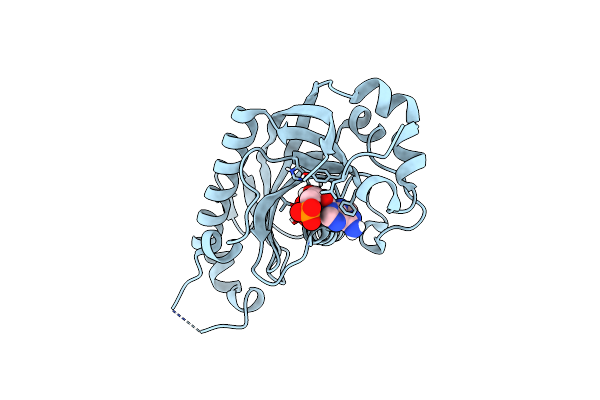 |
Catalytic Domain Of Mouse 2',3'-Cyclic Nucleotide 3'- Phosphodiesterase, With Mutation V321A, Complexed With 2',5'-Adp
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-09-23 Classification: HYDROLASE Ligands: A2P |
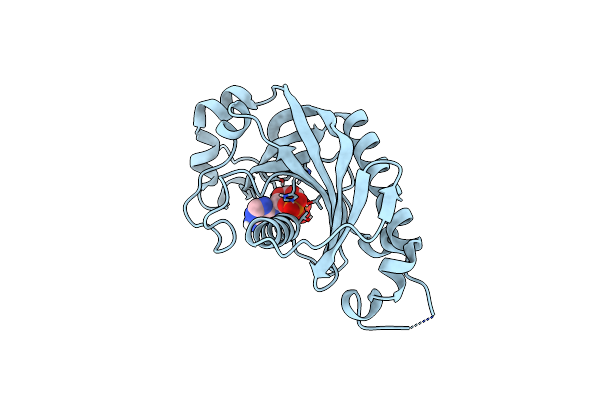 |
Catalytic Domain Of Mouse 2',3'-Cyclic Nucleotide 3'- Phosphodiesterase, With Mutation V321A, Complexed With 2',5'-Adp
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2015-09-23 Classification: HYDROLASE Ligands: A2P |
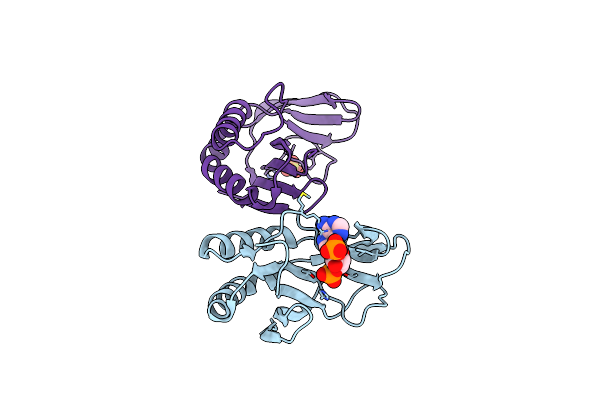 |
The 1.35 Structure Of A Viral Rnase L Antagonist Reveals Basis For The 2'-5'-Oligoadenylate Binding And Enzyme Activity.
Organism: Rotavirus a
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2015-04-29 Classification: VIRAL PROTEIN Ligands: A2P, SO4 |
 |
3D Structure Of E. Coli Isocitrate Dehydrogenase K100M Mutant In Complex With Alpha-Ketoglutarate, Calcium(Ii) And Adenine Nucleotide Phosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-10-31 Classification: OXIDOREDUCTASE Ligands: A2P, AKG, CA, SO4 |
 |
3D Structure Of E. Coli Isocitrate Dehydrogenase K100M Mutant In Complex With Isocitrate, Magnesium(Ii), Adenosine 2',5'-Biphosphate And Ribosylnicotinamide-5'-Phosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-10-31 Classification: OXIDOREDUCTASE Ligands: A2P, ICT, MG, NMN, SO4 |
 |
Crystal Structure Of Aspartate Beta-Semialdehide Dehydrogenase From Streptococcus Pneumoniae With 2',5'-Adenosine Diphosphate And D-2-Aminoadipate
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-01-04 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: 11C, NA, A2P |
 |
Crystal Structure Of The H286L Mutant Of Ferredoxin-Nadp+ Reductase From Plasmodium Falciparum With 2'P-Amp
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-01-12 Classification: OXIDOREDUCTASE Ligands: FAD, A2P |
 |
Crystal Structure Of The H286K Mutant Of Ferredoxin-Nadp+ Reductase From Plasmodium Falciparum In Complex With 2'P-Amp
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-01-12 Classification: OXIDOREDUCTASE Ligands: FAD, A2P |
 |
X-Ray Structure Of The Ferredoxin-Nadp(H) Reductase From Rhodobacter Capsulatus In Complex With 2P-Amp At 2.37 Angstroms Resolution
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2008-11-11 Classification: OXIDOREDUCTASE Ligands: FAD, A2P, HTG |
 |
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2007-01-23 Classification: OXIDOREDUCTASE Ligands: NAP, 5RP, A2P |
 |
Structure Of Aspartate Semialdehyde Dehydrogenase (Asadh) From Streptococcus Pneumoniae Complexed With 2',5'-Adp
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-08-15 Classification: OXIDOREDUCTASE Ligands: A2P |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2003-12-09 Classification: HYDROLASE Ligands: A2P |
 |
Crystal Structure Of Eosinophil Cationic Protein In Complex With 2',5'-Adp At 2.0 A Resolution Reveals The Details Of The Ribonucleolytic Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-10-03 Classification: HYDROLASE Ligands: A2P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-05-31 Classification: HYDROLASE Ligands: A2P |
 |
Refined Crystal Structure Of Spinach Ferredoxin Reductase At 1.7 Angstroms Resolution: Oxidized, Reduced, And 2'-Phospho-5'-Amp Bound States
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1995-04-20 Classification: OXIDOREDUCTASE (NADP+(A),FERREDOXIN(A)) Ligands: SO4, FDA, A2P |

