Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
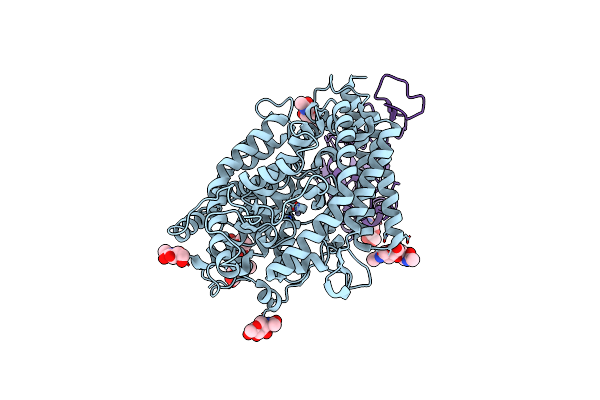
Cryo-Em Structure Of Sars-Cov-2 Omicron Ba.1 Spike Protein In Complex With White-Tailed Deer Ace2
Organism: Severe acute respiratory syndrome coronavirus 2, Human immunodeficiency virus 1, Odocoileus virginianus texanus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRAL PROTEIN Ligands: NAG, ZN |
 |
Sars-Cov-2 Omicron Ba.1 Spike Protein Receptor-Binding Domain In Complex With White-Tailed Deer Ace2
Organism: Severe acute respiratory syndrome coronavirus 2, Odocoileus virginianus texanus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRAL PROTEIN Ligands: NAG, ZN |
 |
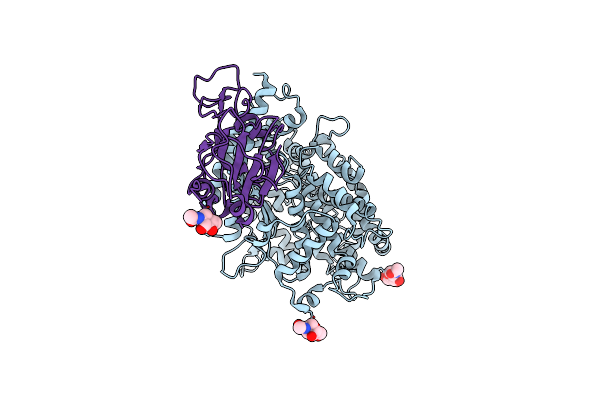
Cryo-Em Structure Of Sars-Cov-2 Prototype Spike Protein In Complex With White-Tailed Deer Ace2
Organism: Severe acute respiratory syndrome coronavirus 2, Odocoileus virginianus texanus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRAL PROTEIN Ligands: NAG, ZN |
 |
Cryo-Em Structure Of Sars-Cov-2 Prototype Spike Protein Receptor-Binding Domain In Complex With White-Tailed Deer Ace2
Organism: Odocoileus virginianus texanus, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRAL PROTEIN Ligands: NAG, ZN |
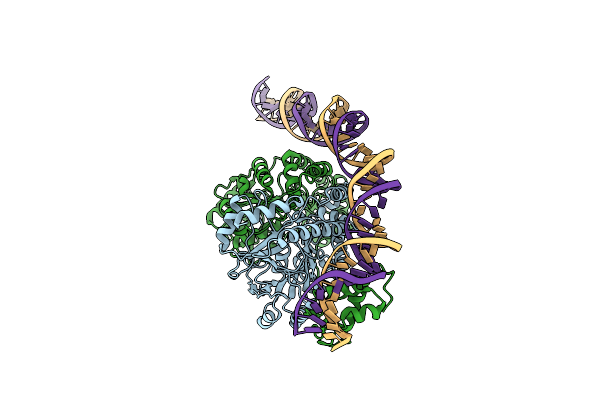 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Half-Closed Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
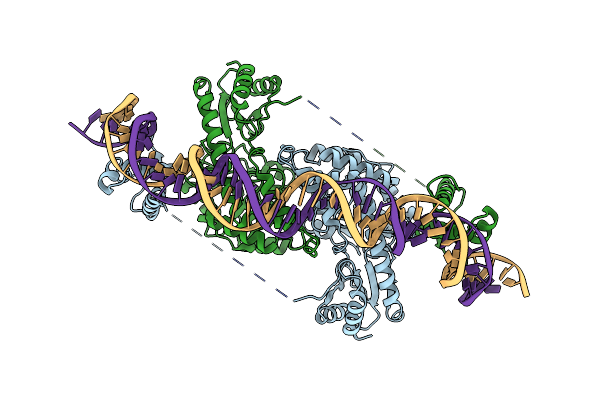 |
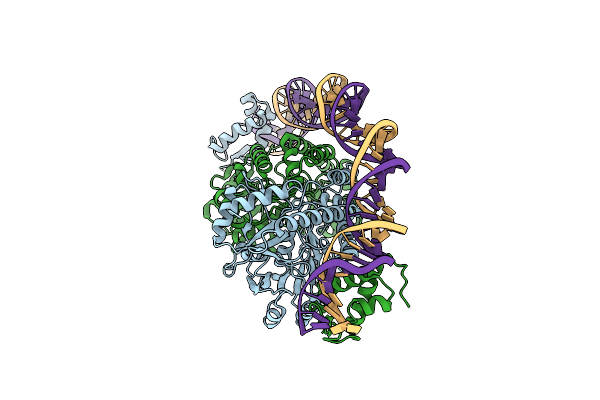
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C2 Symmetry.
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
 |
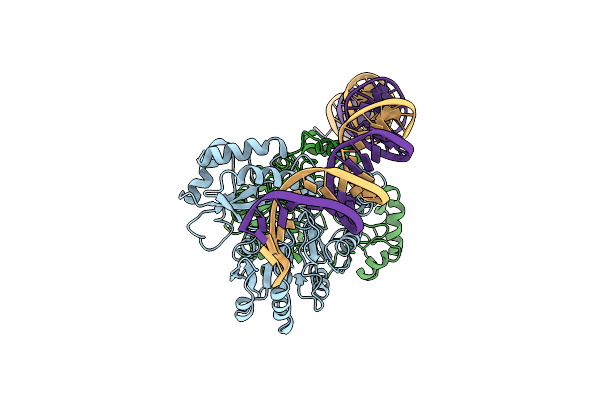
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C1 Symmetry
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Open Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Alkalihalobacillus clausii
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-03-29 Classification: LYASE Ligands: PEG, CA |
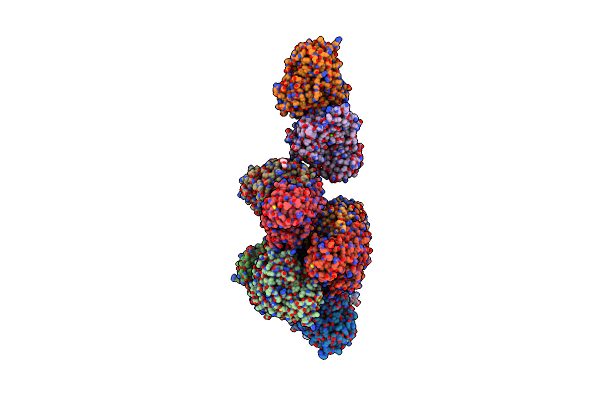 |
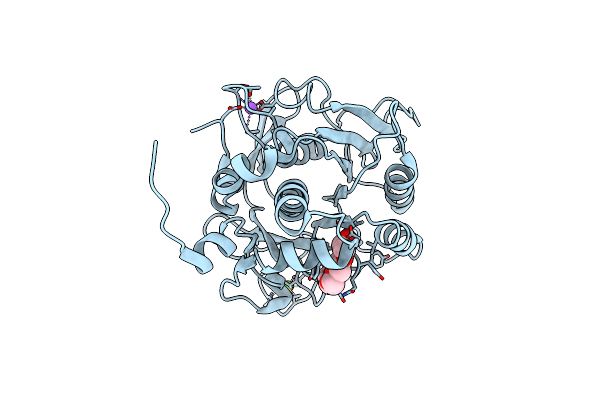
Organism: Alkalihalobacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-09-28 Classification: DNA BINDING PROTEIN Ligands: CA, CL, PGE, EDO, PEG, P6G |
 |
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-03-16 Classification: HYDROLASE |
 |
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2010-09-08 Classification: HYDROLASE |
 |
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2010-09-08 Classification: HYDROLASE |
 |
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2010-08-11 Classification: HYDROLASE |
 |
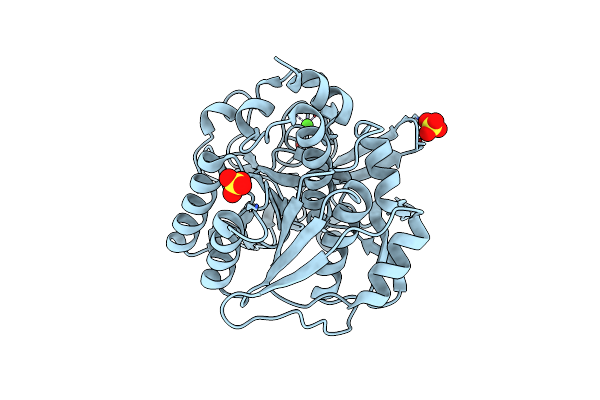
High Resolution Crystal Structure Of The Intact Pro-Kumamolisin, A Sedolisin Type Proteinase (Previously Called Kumamolysin Or Kscp)
Organism: Bacillus sp. mn-32
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: CA |
 |
High Resolution Crystal Structure Of Mutant E23A Of Kumamolisin, A Sedolisin Type Proteinase (Previously Called Kumamolysin Or Kscp)
Organism: Bacillus sp. mn-32
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: CA, SO4 |
 |
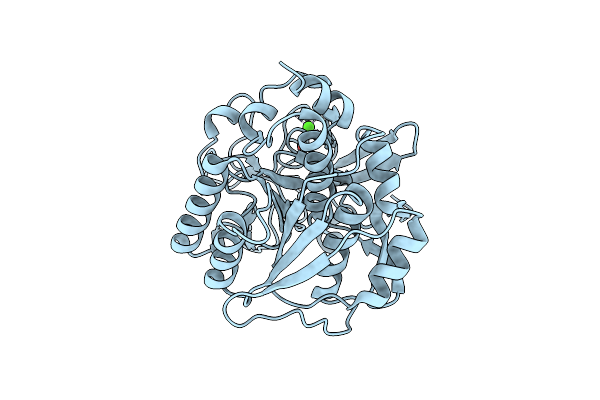
High Resolution Crystal Structure Of Mutant W129A Of Kumamolisin, A Sedolisin Type Proteinase (Previously Called Kumamolysin Or Kscp)
Organism: Bacillus sp. mn-32
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: CA, SO4 |
 |
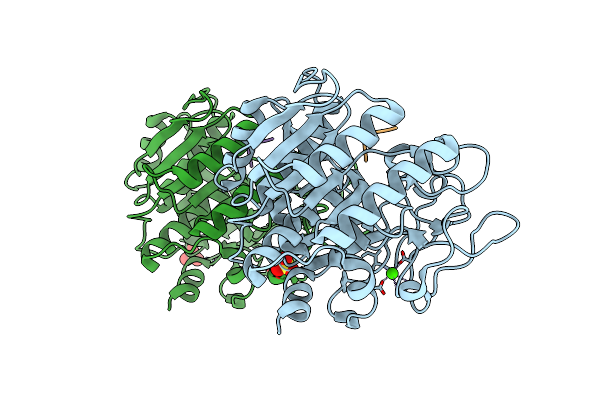
Crystal Structure Of The Thermostable Serine-Carboxyl Type Proteinase, Kumamolysin (Kscp)
Organism: Bacillus novosp. mn-32
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2002-06-13 Classification: HYDROLASE Ligands: CA |
 |
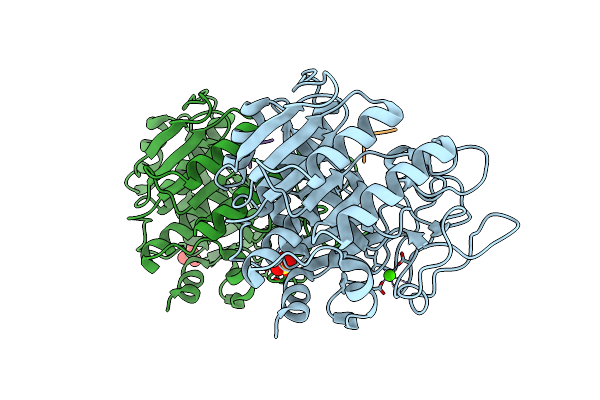
Crystal Structure Of The Thermostable Serine-Carboxyl Type Proteinase, Kumamolisin (Kscp) - Complex With Ac-Ile-Ala-Phe-Cho
Organism: Bacillus novosp. mn-32, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2002-06-13 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, CA |
 |
The Thermostable Serine-Carboxyl Type Proteinase, Kumamolisin (Kscp) - Complex With Ac-Ile-Pro-Phe-Cho
Organism: Bacillus novosp. mn-32, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2002-06-13 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, SO4 |