Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
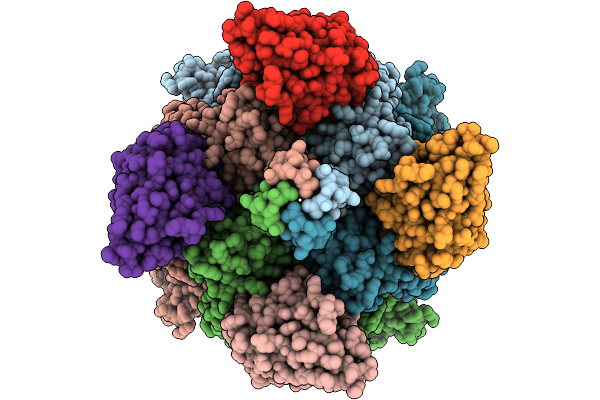 |
Organism: Camelus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, CA, A1L5I |
 |
Organism: Homo sapiens, Camelus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: ZN, CA, A1L5I |
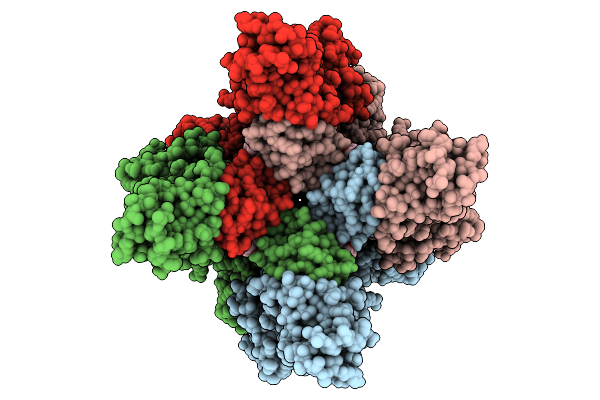 |
Organism: Homo sapiens, Camelus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: METAL TRANSPORT Ligands: CA, ZN, A1L5I |
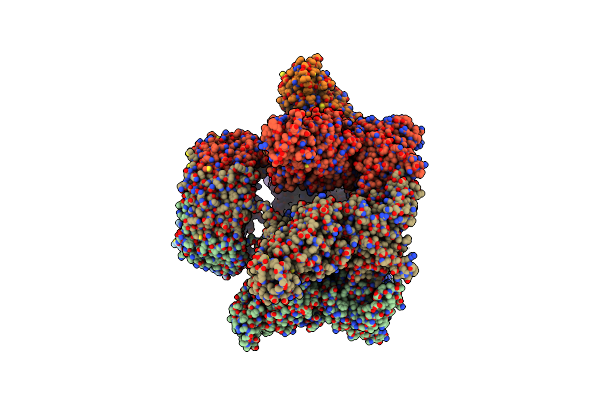 |
Trwk/Virb4Unbound Trimer Of Dimers Complex (With Hcp1) From The R388 Type Iv Secretion System Determined By Cryo-Em.
Organism: Salmonella dublin, Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: ELECTRON MICROSCOPY Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN |
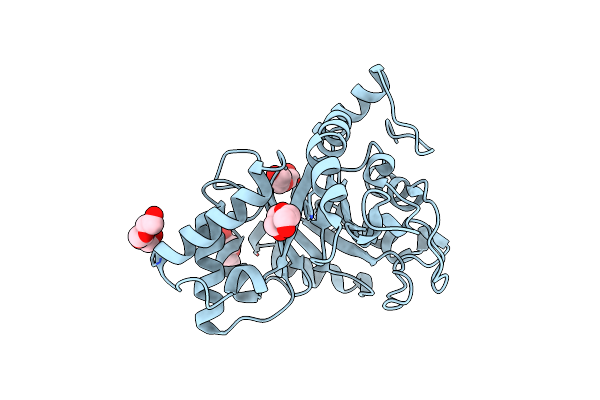 |
Crystal Structure Of Endo-Beta-N-Acetylglucosaminidase From Cordyceps Militaris In Apo Form
Organism: Cordyceps militaris cm01
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: PGE, PEG |
 |
Crystal Structure Of Endo-Beta-N-Acetylglucosaminidase From Cordyceps Militaris In Complex With L-Fucose
Organism: Cordyceps militaris cm01
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: FUC, PGE, PEG |
 |
Crystal Structure Of Endo-Beta-N-Acetylglucosaminidase From Cordyceps Militaris D154N/E156Q Mutant In Complex With Fucosyl-N-Acetylglucosamine
Organism: Cordyceps militaris cm01
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-02 Classification: HYDROLASE |
 |
Crystal Structure Of Endo-Beta-N-Acetylglucosaminidase From Cordyceps Militaris D154N/E156Q Mutant In Complex With Fucosyl-N-Acetylglucosamine-Asn
Organism: Cordyceps militaris cm01
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: PGE, PEG, ASN |
 |
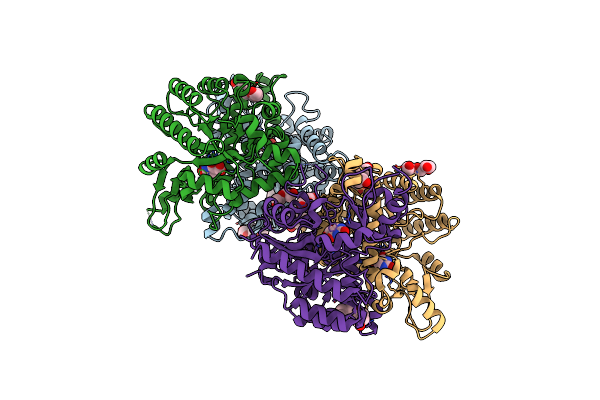
Organism: Cordyceps militaris
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2013-10-02 Classification: LYASE Ligands: ZN |
 |
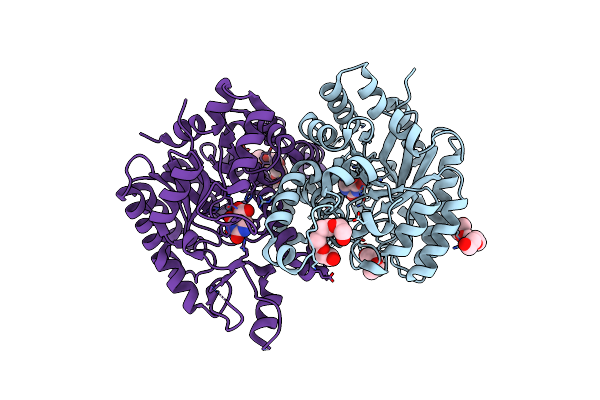
Crystal Structure Of Cordyceps Militaris Idcase D323A Mutant In Complex With 5-Carboxyl-Uracil
Organism: Cordyceps militaris
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-10-02 Classification: LYASE Ligands: ZN, 5CU, P6G |
 |
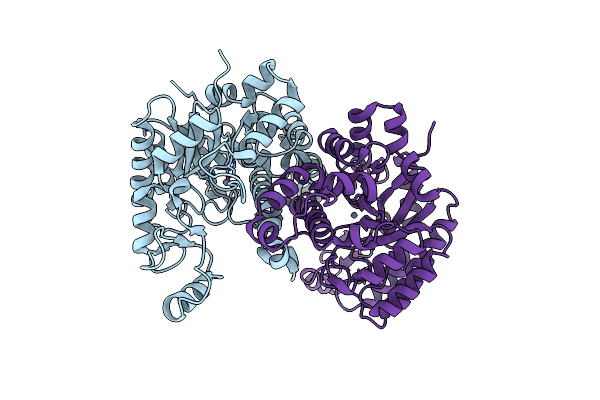
Crystal Structure Of Cordyceps Militaris Idcase D323N Mutant In Complex With 5-Carboxyl-Uracil
Organism: Cordyceps militaris
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-10-02 Classification: LYASE Ligands: ZN, 5CU, P6G |
 |
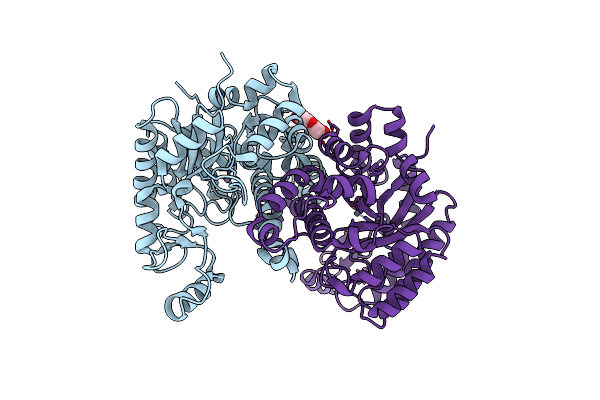
Organism: Cordyceps militaris
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-10-02 Classification: LYASE Ligands: ZN |
 |
Organism: Cordyceps militaris
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-10-02 Classification: LYASE Ligands: ZN, PEG |
 |
Crystal Structure Of The N-Terminal Fragment Of Zebrafish Tlr5 In Complex With Salmonella Flagellin
Organism: Danio rerio, Eptatretus burgeri, Salmonella enterica subsp. enterica serovar dublin
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2012-02-29 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Salmonella enterica subsp. enterica serovar dublin
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2006-05-16 Classification: HYDROLASE Ligands: SO4 |
 |
 |
 |
 |
 |
NA
Organism: Cordyceps militaris (strain CM01)
Method: Alphafold Release Date: Classification: NA Ligands: NA |