Search Count: 1,394
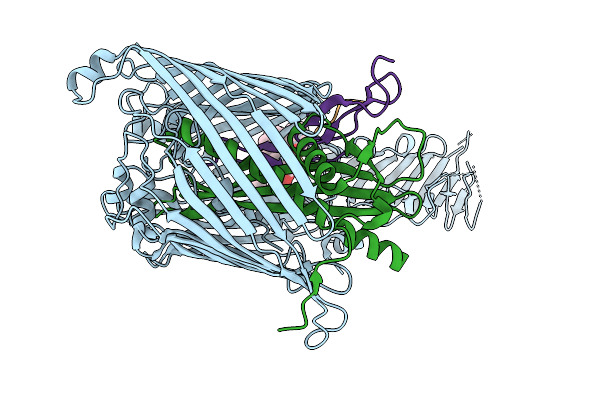 |
Cryo-Em Structure Of Shigella Flexneri Lptde In Complex With Rtp45 Superinfection Exclusion Protein From Rtp Bacteriophage And Endogenous Lptm
Organism: Shigella flexneri, Escherichia phage rtp, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
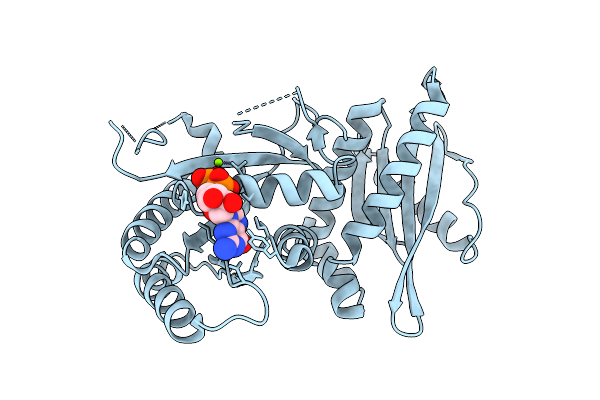 |
Crystal Structure Of Monomeric Rag-Like Small Gtpase From Asgard Lokiarchaeota (Lokiragm) In Complex With Gdp
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: HYDROLASE Ligands: GDP, MG |
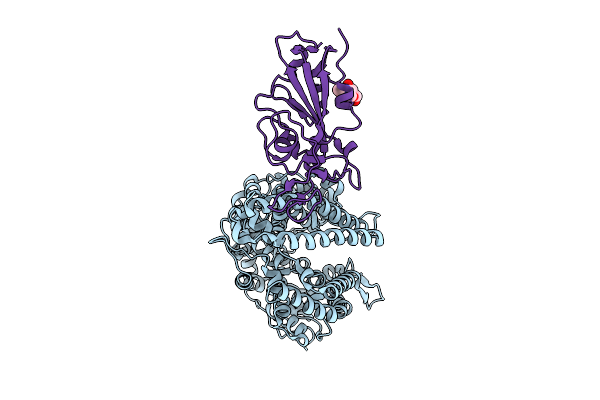 |
Organism: Ovis aries, Bat coronavirus ratg13
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
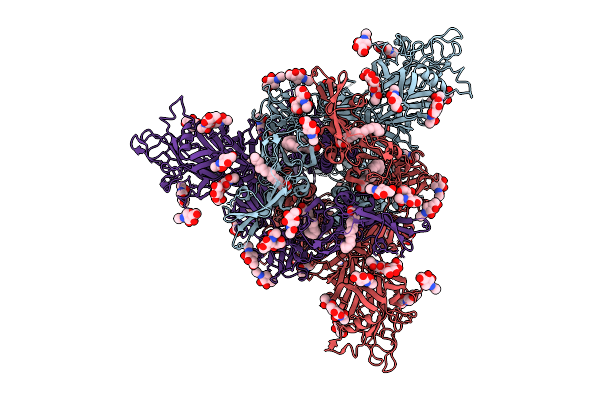 |
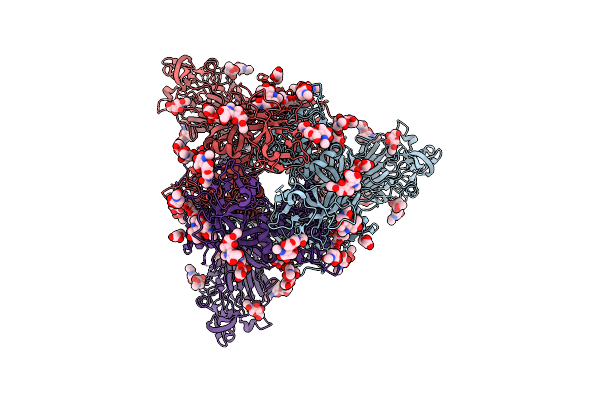
Organism: Bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: PLM, NAG, OLA |
 |
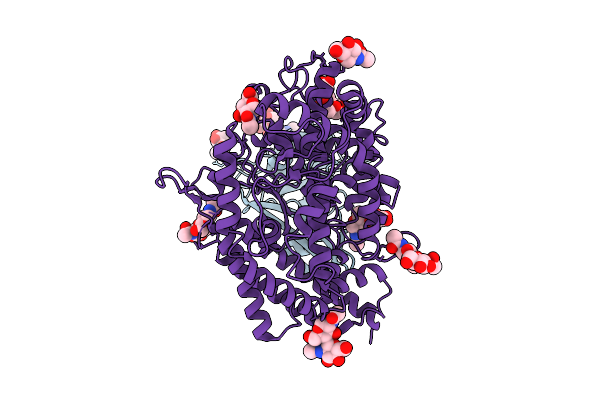
Organism: Bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: PLM, NAG |
 |
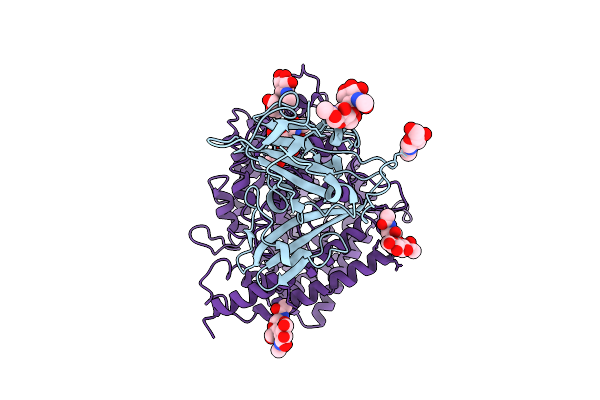
Organism: Bat coronavirus hku5, Pipistrellus abramus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
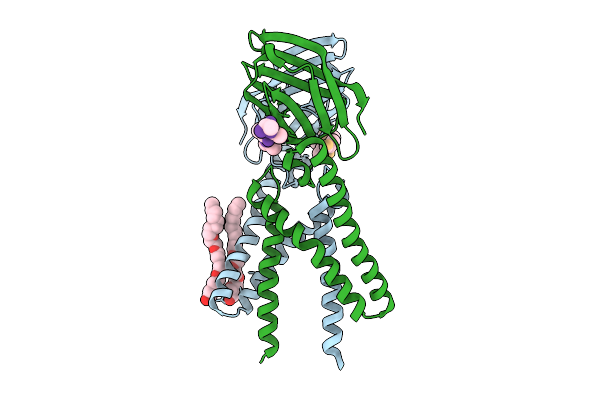
Organism: Bat coronavirus hku5, Pitta sordida
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of A Coronaviral M Protein In Complex With A C-Terminal Peptide Of The N Protein
Organism: Pipistrellus bat coronavirus hku5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: N8E |
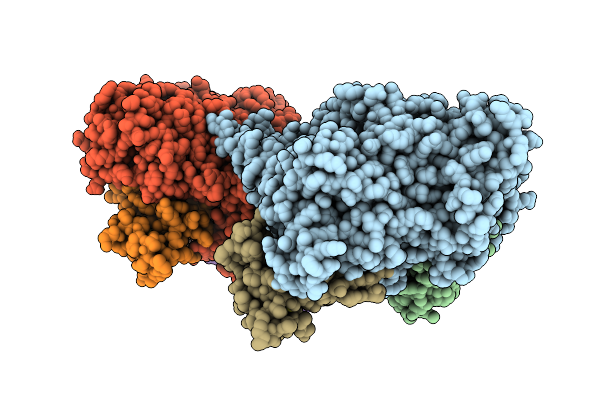 |
Benzylsuccinate Synthase Alpha-Beta-Gamma Complex With Bound Toluene And Fumarate
Organism: Thauera aromatica
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LYASE Ligands: FUM, MBN, PEG, SF4 |
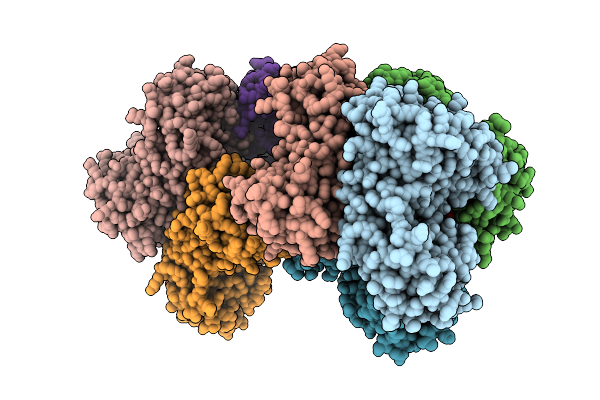 |
Crystal Structure Of Homo Tetrameric Raga-Like Small Gtpase From Asgard Lokiarchaeota (Lokirago) In Complex With Gtp
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE Ligands: GTP, MG |
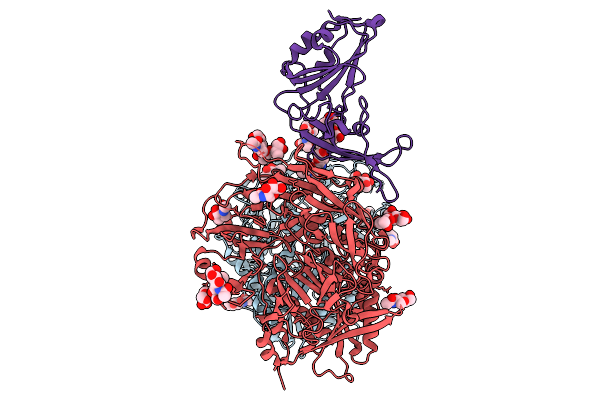 |
Cryo-Em Structure Of Mjhku4R-Cov-1 Receptor-Binding Domain Complexed With Human Cd26
Organism: Homo sapiens, Tylonycteris bat coronavirus hku4
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
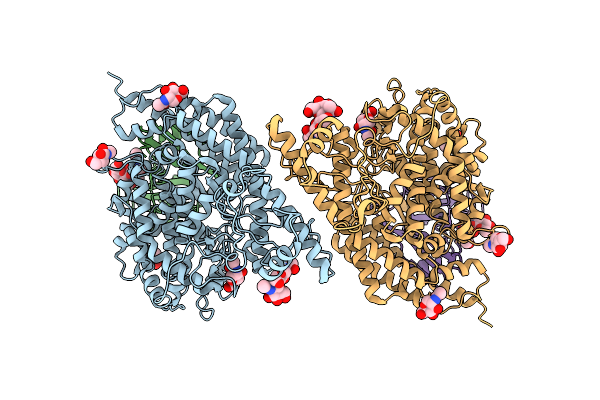 |
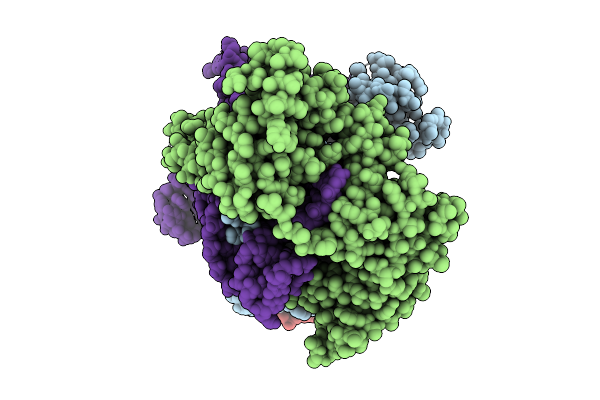
Structure Of Hku5 Spike C-Terminal Domain In Complex With Ace2 From Pipistrellus Abramus
Organism: Pipistrellus abramus, Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Syntrophomonas palmitatica jcm 14374, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
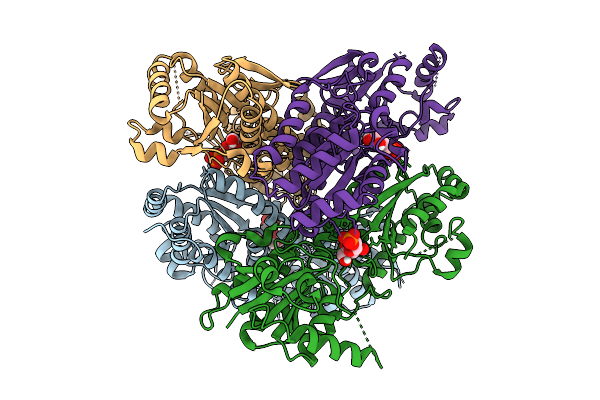
Crystal Structure Of Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE |
 |
Crystal Structure Of The Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum With Fructose 6-Phosphate
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: F6P |
 |
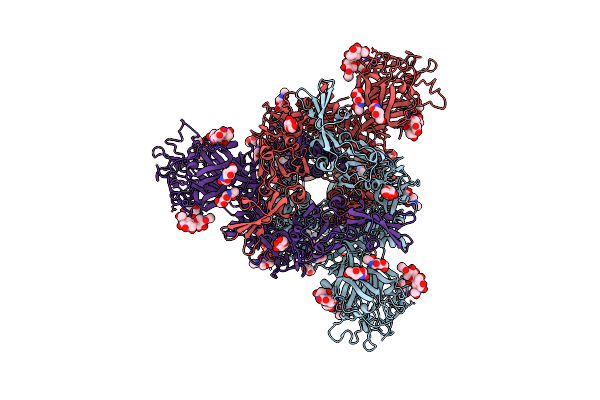
Crystal Structure Of Pyrophosphate-Dependent Phosphofructokinase From Candidatus Prometheoarchaeum Syntrophicum With Phosphoenolpyruvate, Phosphate And Mg2+
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: TRANSFERASE Ligands: PEP, PO4, MG |
 |
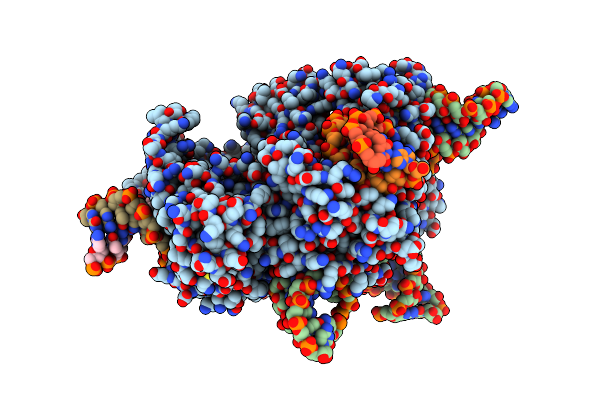
Organism: Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryoem Structure Of Thermocas9 In Post-Cleavage State With A Dna Containing Nnnncga Pam
Organism: Geobacillus thermodenitrificans, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: HYDROLASE/DNA/RNA Ligands: GTP, MG |
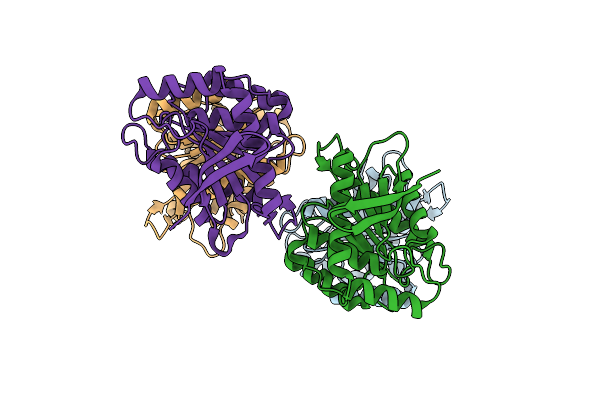 |
Organism: Hydrogenobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-05-14 Classification: HYDROLASE |
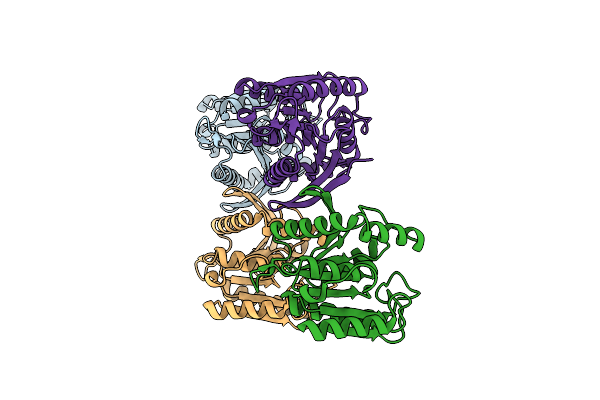 |
Crystal Structure Of Thermostable Dienelactone Hydrolase. Monoclinic Space Group.
Organism: Hydrogenobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-05-14 Classification: HYDROLASE |

