Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
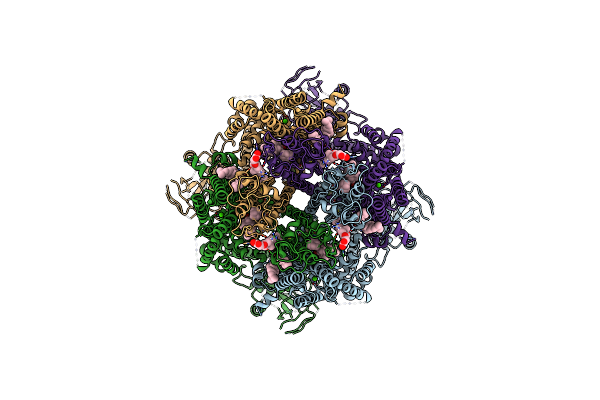 |
Cryo-Em Structure Of The Avian Great Tit Trpm8 Channel In Complex With The Antagonist Tc-I 2014
Organism: Parus major
Method: ELECTRON MICROSCOPY Resolution:3.26 Å Release Date: 2024-08-21 Classification: MEMBRANE PROTEIN Ligands: CA, Y01, T14 |
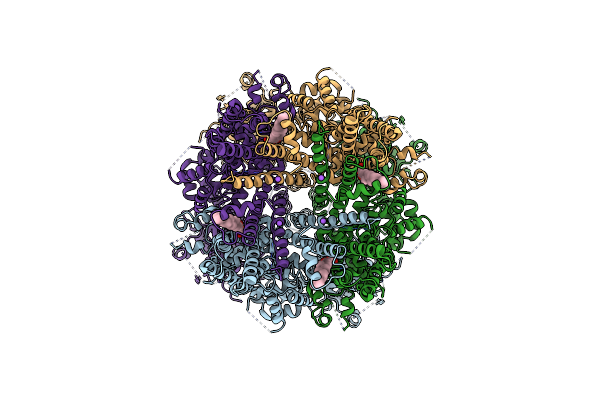 |
Structure Of The Trpm8 Cold Receptor By Single Particle Electron Cryo-Microscopy, Ligand-Free State
Organism: Parus major
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: TRANSPORT PROTEIN Ligands: Y01, NA |
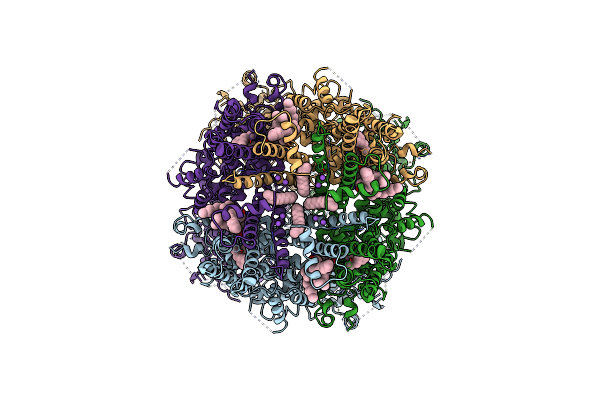 |
Structure Of The Trpm8 Cold Receptor By Single Particle Electron Cryo-Microscopy, Amtb-Bound State
Organism: Parus major
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: TRANSPORT PROTEIN Ligands: Y01, UND, 9PE, LQ7, NA |
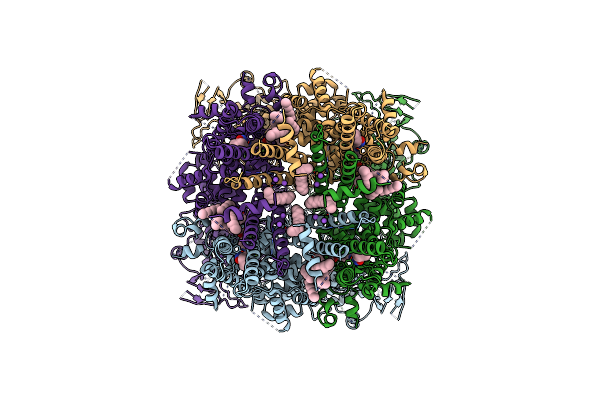 |
Structure Of The Trpm8 Cold Receptor By Single Particle Electron Cryo-Microscopy, Tc-I 2014-Bound State
Organism: Parus major
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: TRANSPORT PROTEIN Ligands: Y01, UND, 9PE, T14, NA |
 |
Structure Of The Trpm8 Cold Receptor By Single Particle Electron Cryo-Microscopy, Calcium-Bound State
Organism: Parus major
Method: ELECTRON MICROSCOPY Release Date: 2019-09-18 Classification: TRANSPORT PROTEIN Ligands: CA, Y01 |
 |
Crystal Structure Of Designed Two-Component Self-Assembling Icosahedral Cage I53-40
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440), Vibrionales bacterium (strain swat-3)
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2016-07-27 Classification: PROTEIN BINDING |
 |
Crystal Structure Of The Mutant P311A Of Enolase Superfamily Member From Vibrionales Bacterium Complexed With Mg And D-Arabinonate
Organism: Vibrionales bacterium swat-3
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-06-06 Classification: ISOMERASE Ligands: MG, EPE, D8T |
 |
Crystal Structure Of Probable Keto-Hydroxyglutarate-Aldolase From Vibrionales Bacterium Swat-3 (Target Efi-502156)
Organism: Vibrionales bacterium swat-3
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2012-03-21 Classification: LYASE Ligands: CL |
 |
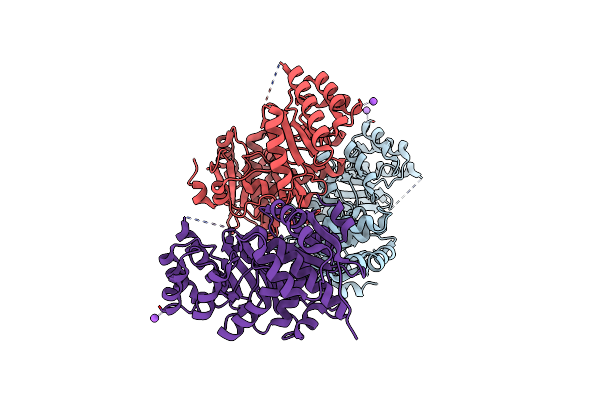
Crystal Structure Of Enolase Superfamily Member From Vibrionales Bacterium Complexed With Mg And Glycerol In The Active Site
Organism: Vibrionales bacterium swat-3
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-03-14 Classification: ISOMERASE Ligands: MG, GOL |
 |
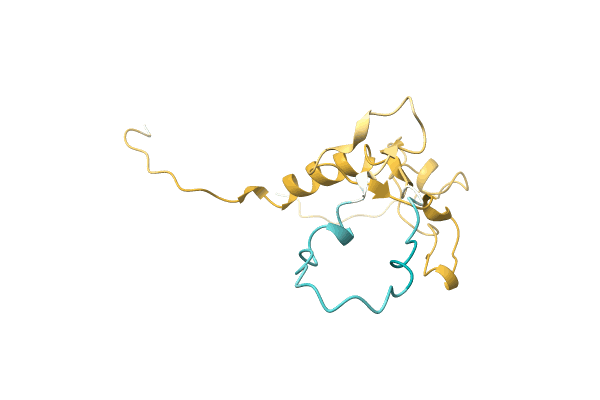
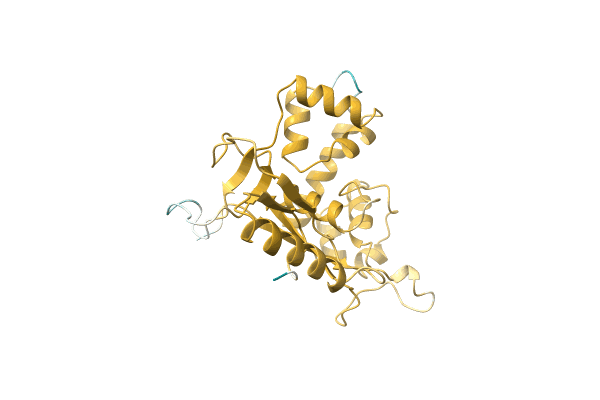
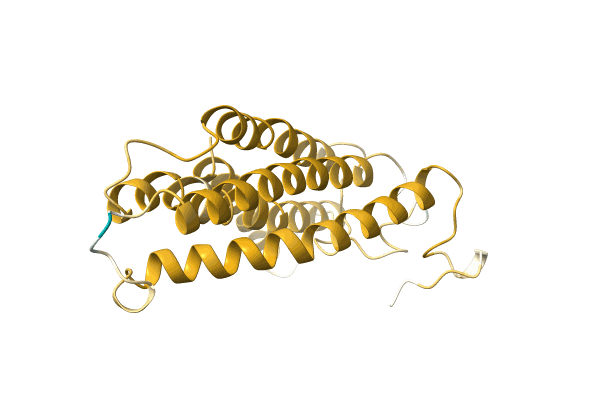
Crystal Structure Of Putative Mandelate Racemase / Muconate Lactonizing Enzyme From Vibrionales Bacterium Swat-3
Organism: Vibrionales bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-07-01 Classification: ISOMERASE |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |