Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Crassostrea gigas
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: ANTIVIRAL PROTEIN Ligands: C2E |
 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: CHAPERONE Ligands: MG, ADP, AF3, K |
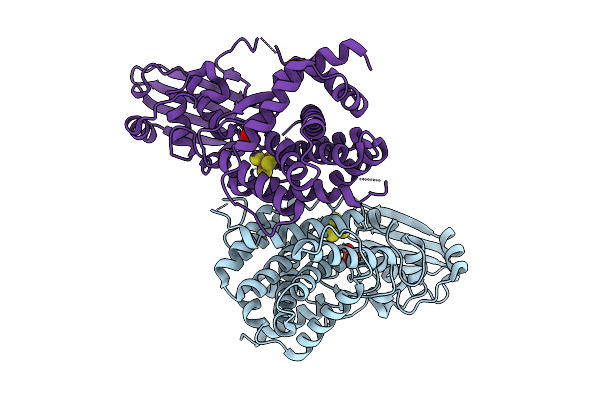 |
[4Fe-4S] Cluster-Containing L-Cysteine Desulfidase Cyua From Methanococcus Maripaludis
Organism: Methanococcus maripaludis
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: SF4, SO4 |
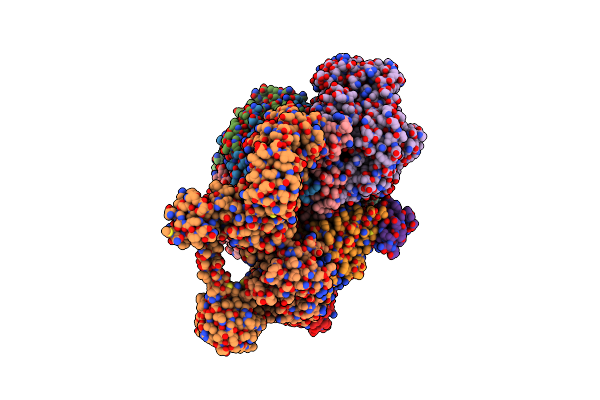 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: SHT, TP7, COM, F43, S5Q, ZN, ATP, MG |
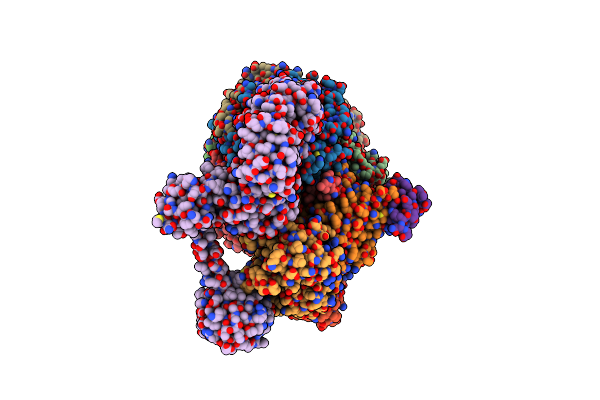 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, SHT, COM, TP7, S5Q |
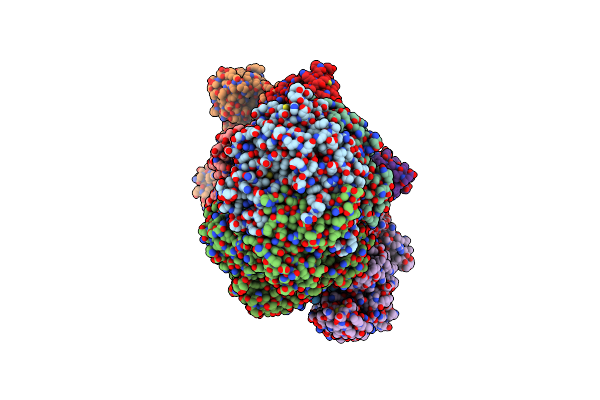 |
Methyl-Coenzyme M Reductase Activation Complex Binding To The A2 Component After Incubation With Atp
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, COM, TP7, SHT, S5Q, ZN, ATP, MG |
 |
Cryo-Em Structure Of The Avian Great Tit Trpm8 Channel In Complex With The Antagonist Tc-I 2014
Organism: Parus major
Method: ELECTRON MICROSCOPY Resolution:3.26 Å Release Date: 2024-08-21 Classification: MEMBRANE PROTEIN Ligands: CA, Y01, T14 |
 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-08-14 Classification: LYASE Ligands: GOL |
 |
Crystal Structure Of 3-Ketosteroid Delta1-Dehydrogenase From Rhodococcus Erythropolis Sq1 In Complex With 1,4-Androstadiene-3,17- Dione
Organism: Rhodococcus electrodiphilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-26 Classification: OXIDOREDUCTASE Ligands: FAD, ANB |
 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-04-03 Classification: TRANSFERASE |
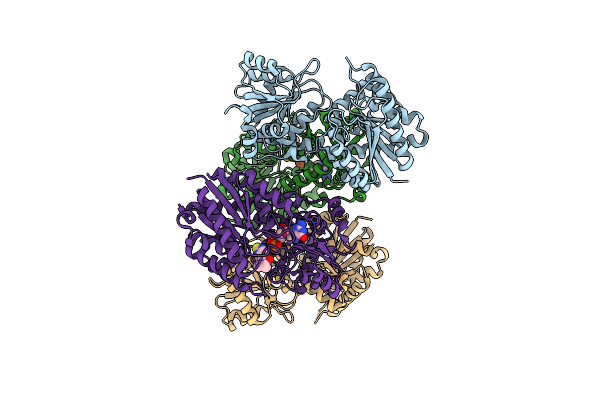 |
Structure Of Glycosyltransferase Lmbt In Complex With Gdp And Ergothioneine
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-04-03 Classification: TRANSFERASE Ligands: LW8, GDP |
 |
Organism: Methanococcus maripaludis
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: SF4, IOD, CL |
 |
Organism: Methanococcus maripaludis
Method: X-RAY DIFFRACTION Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: SF4, AMP, CL |
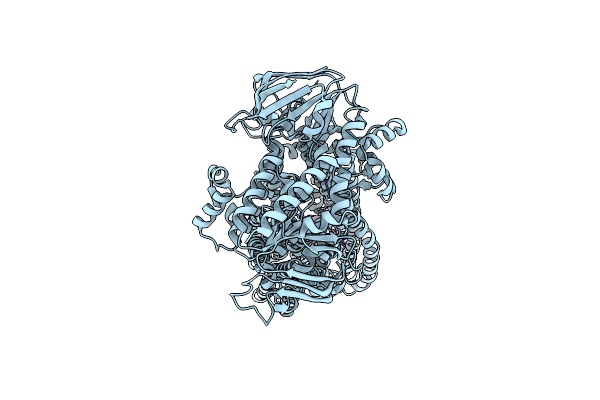 |
Bovine Multidrug Resistance Protein 4 (Mrp4) Bound To Prostaglandin E2 In Msp Lipid Nanodisc
Organism: Bos indicus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSPORT PROTEIN Ligands: P2E |
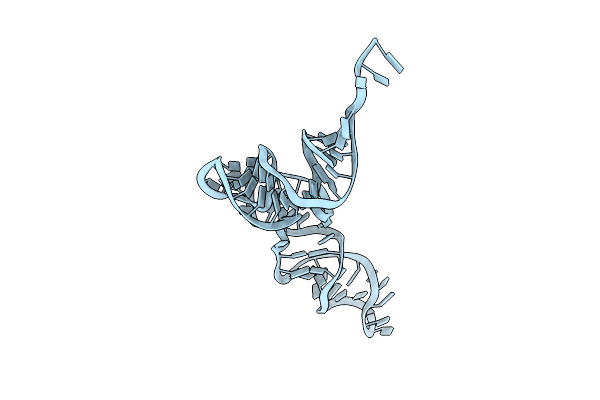 |
Organism: Candidatus methanomethylophilus alvus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: RNA |
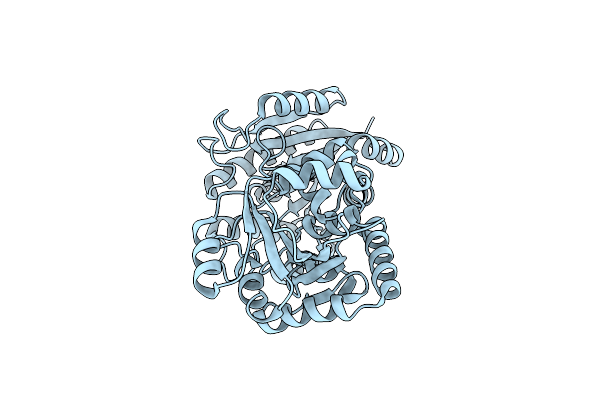 |
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2023-09-20 Classification: BIOSYNTHETIC PROTEIN |
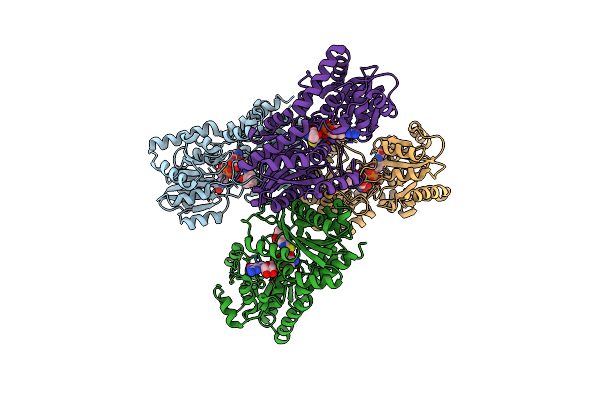 |
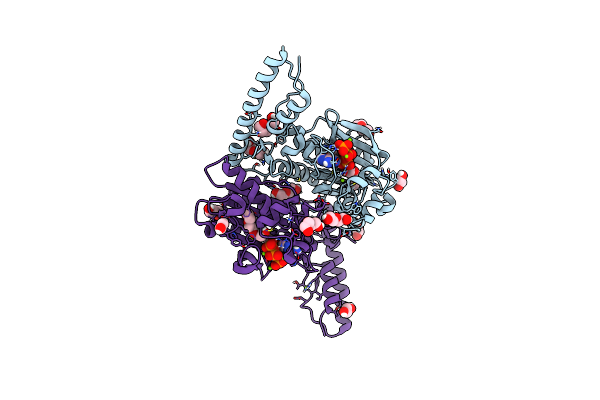
Crystal Structure Of Lmbt From Streptomyces Lincolnensis Nrrl Isp-5355 In Complex With Substrates
Organism: Streptomyces lincolnensis
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2023-09-20 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, Q3L |
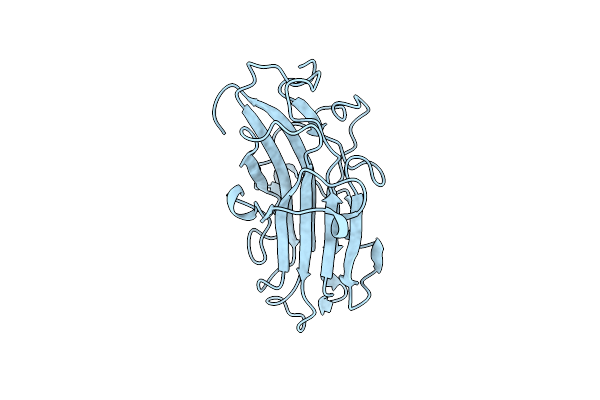 |
Organism: Polaribacter haliotis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-09-20 Classification: SUGAR BINDING PROTEIN |
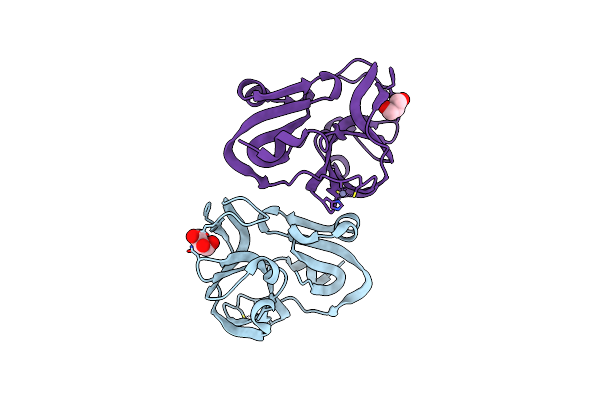 |
Organism: Human coxsackievirus a16 (strain g-10)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-08-02 Classification: VIRAL PROTEIN Ligands: GOL, ZN |
 |
Crystal Structure Of Pyrrolysyl-Trna Synthetase From Methanomethylophilus Alvus Engineered For 3-Methyl-L-Histidine, Bound To Amppnp
Organism: Candidatus methanomethylophilus alvus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-07-19 Classification: LIGASE Ligands: ANP, PEG, PGE, EDO, MG |