Search Count: 7
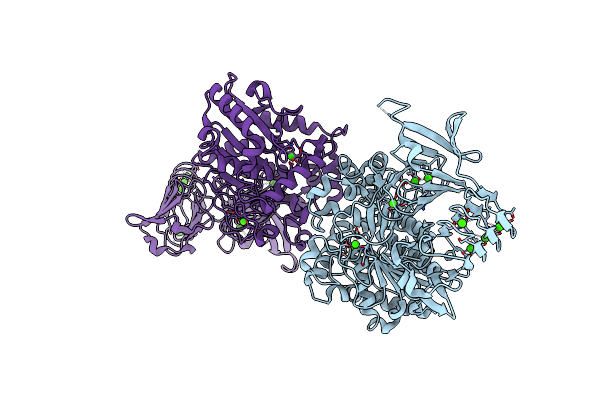 |
Crystal Structure Of Pseudomonas Sp. Mis38 Lipase (Pml) In The Open Conformation Following Dialysis Against Ca-Free Buffer
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2010-05-26 Classification: HYDROLASE Ligands: CA |
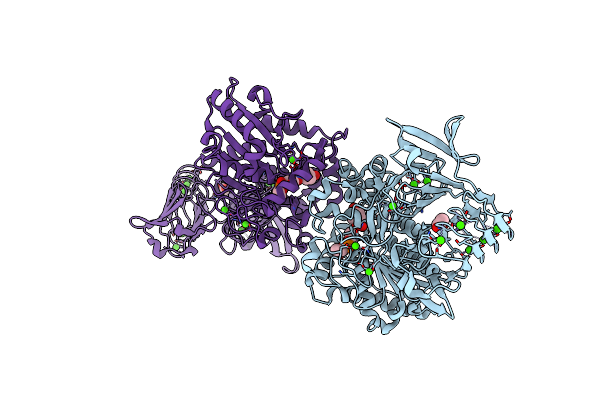 |
Crystal Structure Of Pseudomonas Sp. Mis38 Lipase In Complex With Diethyl Phosphate
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2010-05-26 Classification: HYDROLASE Ligands: CA, DEP, NPO, ACT |
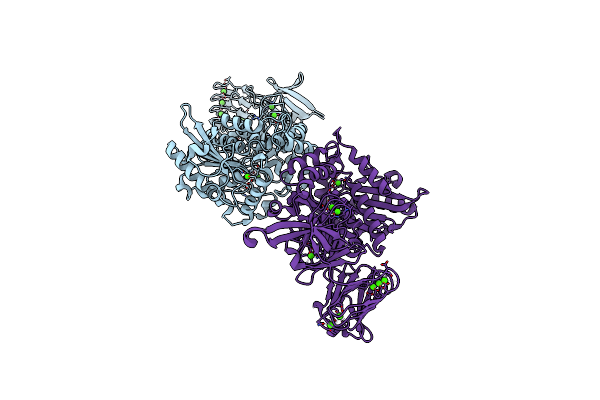 |
Organism: Pseudomonas
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2009-11-10 Classification: HYDROLASE Ligands: CA |
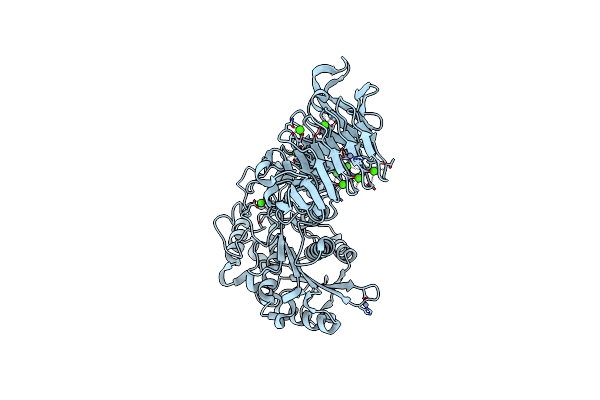 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2008-12-02 Classification: HYDROLASE Ligands: CA, ZN |
 |
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2008-12-02 Classification: HYDROLASE Ligands: CA, ZN |
 |
Organism: Pseudomonas sp. mis38
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2007-10-30 Classification: HYDROLASE Ligands: CA, ZN |
 |
Crystal Structure Of A Platinum-Bound S445C Mutant Of Pseudomonas Sp. Mis38 Lipase
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-10-30 Classification: HYDROLASE Ligands: CA, ZN, PT |

