Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
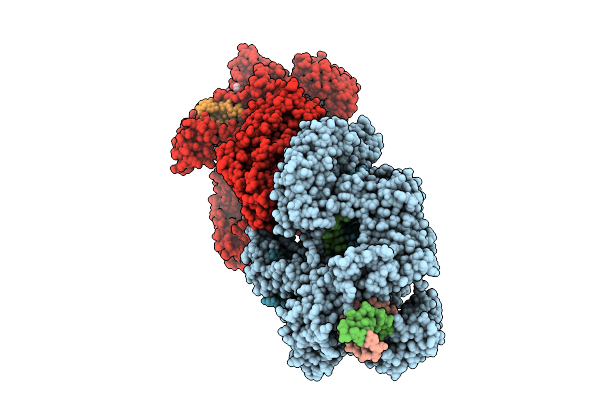 |
Organism: Acidaminococcus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: SPLICING/DNA/RNA |
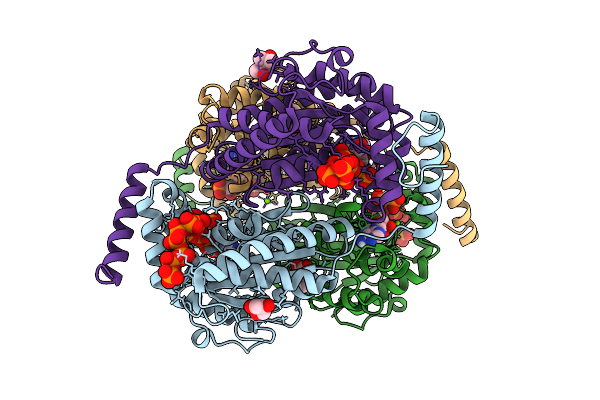 |
Crystal Structure Of Ppk2 Class Iii From Erysipelotrichaceae Bacterium In Complex With Appch2P And Polyphosphate
Organism: Erysipelotrichaceae bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: ACP, 6YY, GOL, BEZ, MG, MPD, A1I4D |
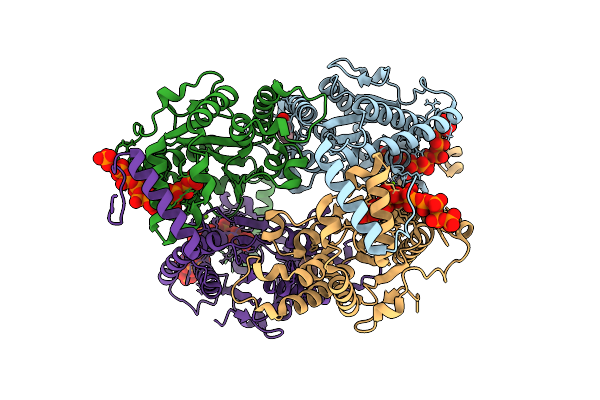 |
Crystal Structure Of Ppk2 Class Iii From Erysipelotrichaceae Bacterium In Complex With Polyphosphate
Organism: Erysipelotrichaceae bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: 6YY, BEZ, PO4, MG |
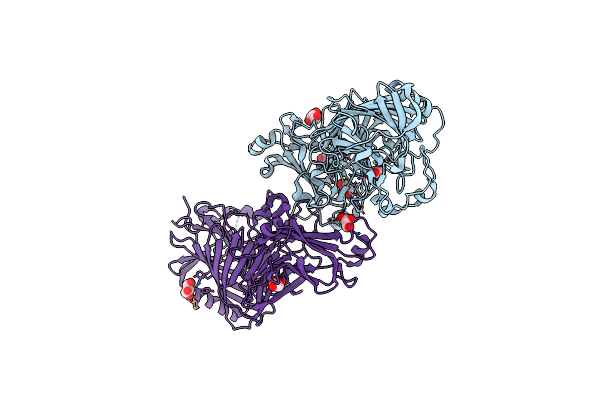 |
Xanthomonas Campestris Pv. Campestris Opgd Mutant-D379N With Beta-1,2-Glucan
Organism: Xanthomonas campestris pv. campestris str. atcc 33913
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-11-06 Classification: TRANSFERASE Ligands: GOL |
 |
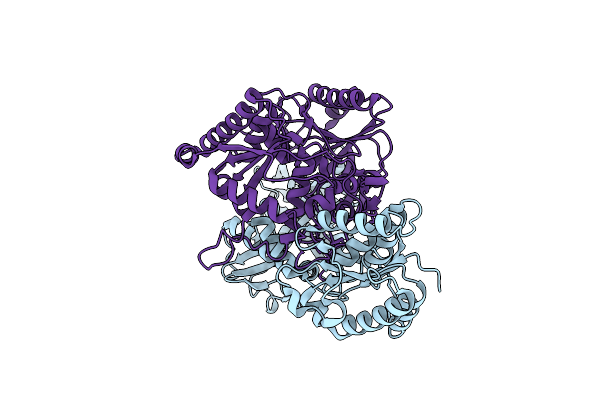
Crystal Structure Of Cis-Epoxysuccinic Hydrolase From Bradyrhizobium Mercantei
Organism: Bradyrhizobium mercantei
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE |
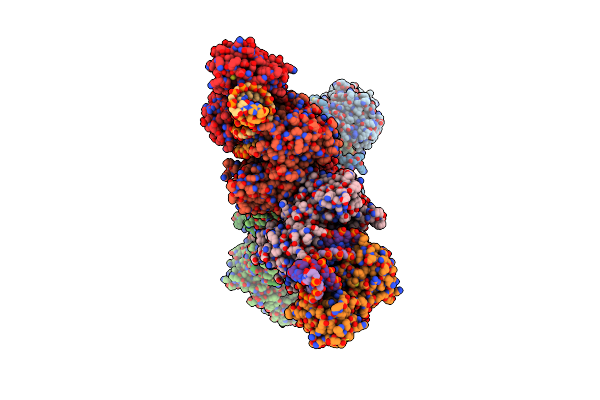
 |
Organism: Bacillales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: IMMUNE SYSTEM |
 |
Cryoem Structure Of The Rna/Dna Bound Sparta (Babago/Tir-Apaz) Tetrameric Complex
Organism: Bacillales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: IMMUNE SYSTEM Ligands: MG |
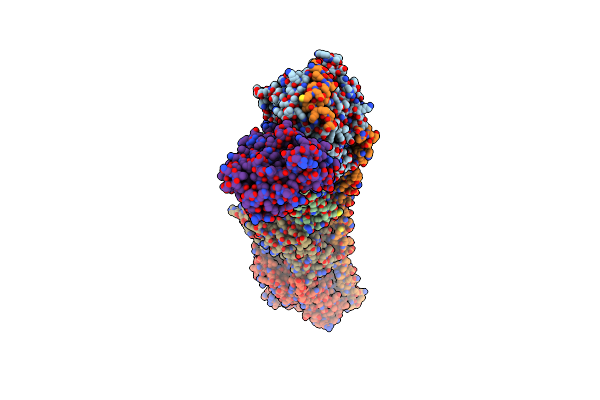 |
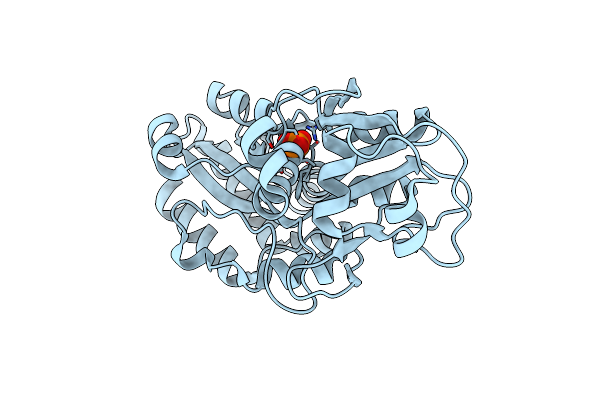
Organism: Mus musculus, Pavo cristatus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, BKF, ACP |
 |
Phosphate-Binding Protein (Psts) From Xanthomonas Citri Pv. Citri A306 Bound To Phosphate
Organism: Xanthomonas citri pv. citri str. 306
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-10-11 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Crystal Structure Of Gh42 Beta-Galactosidase Bibga42A From Bifidobacterium Longum Subspecies Infantis In Complex With Glycerol
Organism: Bifidobacterium longum subsp. infantis (strain atcc 15697 / dsm 20088 / jcm 1222 / nctc 11817 / s12)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-06-07 Classification: HYDROLASE Ligands: GOL, PEG |
 |
Crystal Structure Of Gh42 Beta-Galactosidase Bibga42A From Bifidobacterium Longum Subspecies Infantis E160A/E318A Mutant In Complex With Galactose
Organism: Bifidobacterium longum subsp. infantis (strain atcc 15697 / dsm 20088 / jcm 1222 / nctc 11817 / s12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-07 Classification: HYDROLASE Ligands: GLA |
 |
Crystal Structure Of Gh42 Beta-Galactosidase Bibga42A From Bifidobacterium Longum Subspecies Infantis E318S Mutant In Complex With Lacto-N-Tetraose
Organism: Bifidobacterium longum subsp. infantis (strain atcc 15697 / dsm 20088 / jcm 1222 / nctc 11817 / s12)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-06-07 Classification: HYDROLASE |
 |
Crystal Structure Of Pn-Sia28 In Complex With Influenza Hemagglutinin A/Swine/Guangdong/104/2013 (H1N1)
Organism: Influenza a virus (a/swine/guangdong/104/2013(h1n1)), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-12-21 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of A Glutathione S-Transferase Tau1 From Pinus Densata In Complex With Gsh
Organism: Pinus densata
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-12-07 Classification: TRANSFERASE Ligands: GSH |
 |
Organism: Vibrio sp. g15-21
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: ZN, MG, PO4, EDO, CL, TRS |
 |
Organism: Vibrio sp. g15-21
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: ZN, MG, 15P, CL, EPE, PO4 |
 |
Organism: Vibrio sp. g15-21
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: ZN, MG, EDO, PO4, CL |
 |
Organism: Vibrio sp. g15-21
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: ZN, BR, MG, PO4 |
 |
Wild Type Glta From Bifidobacterium Infantis Jcm 1222 Complexed With Lacto-N-Tetraose
Organism: Bifidobacterium longum subsp. infantis (strain atcc 15697 / dsm 20088 / jcm 1222 / nctc 11817 / s12)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-09-14 Classification: TRANSLOCASE Ligands: SO4 |
 |
Glta N83K Mutant From Bifidobacterium Infantis Jcm 1222 Complexed With Lacto-N-Tetraose
Organism: Bifidobacterium longum subsp. infantis (strain atcc 15697 / dsm 20088 / jcm 1222 / nctc 11817 / s12)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-09-14 Classification: TRANSLOCASE Ligands: SO4, ZN |