Search Count: 112
 |
Structure Of The Chlamydomonas Reinhardtii Respiratory Supercomplex I1 Iii2 Iv2
|
 |
Structure Of The Chlamydomonas Reinhardtii Respiratory Complex I From Respiratory Supercomplex
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: FES, FMN, SF4, PTY, 8Q1, NDP, UQ5, PC7, 3PH, CDL, PGT, COO, ZN |
 |
Subtomogram Average Of The Chlamydomonas Reinhardtii Mitochondrial Respirasome I2 Iii4 Iv6
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: HEM, HEC, ZN, HEA, CU, MG, CUA, FES, FMN, SF4, 8Q1, NDP, COO |
 |
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: FE2, FES, 8Q1 |
 |
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: 8Q1, FE2, FES |
 |
Structure Of The Core Isc Complex Under Turnover Conditions (Frataxin-Bound)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: FE2, 8Q1 |
 |
Structure Of The Core Isc Complex Under Turnover Conditions (Fdx2-Bound In Proximal Conformation)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: 8Q1, FE2, FES |
 |
Structure Of The Core Isc Complex Under Turnover Conditions (Fdx2-Bound In Distal Conformation)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: 8Q1, FE2, FES |
 |
Structure Of Human Cysteine Desulfurase Nfs1 With L-Propargylglycine Bound To Active Site Plp In Complex With Isd11, Acp1 And Iscu2
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-09-11 Classification: TRANSFERASE Ligands: EDO, PLP, LPH, GOL, PEG, PG4, P15, DTT, PGE, EDT, 8Q1, MES, 1PE |
 |
Structure Of The Human Mitochondrial Iron-Sulfur Cluster Biosynthesis Complex During Persulfide Transfer (Persulfide On Iscu2)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: PLP, 8Q1, FE2 |
 |
Structure Of The Human Mitochondrial Iron-Sulfur Cluster Biosynthesis Complex During Persulfide Transfer (Persulfide On Nfs1 And Iscu2)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: PLP, 8Q1, FE2 |
 |
Structure Of The Human Mitochondrial Iron-Sulfur Cluster Biosynthesis Complex During Persulfide Transfer (Without Frataxin)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: PLP, 8Q1, FE2 |
 |
Cryo-Em Structure Of The I-Ii-Iii2-Iv2 Respiratory Supercomplex From Tetrahymena Thermophila
|
 |
Cryo-Em Structure Of The Arabidopsis Thaliana I+Iii2 Supercomplex (Complete Conformation 2 Composition)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: MEMBRANE PROTEIN Ligands: SF4, FES, FMN, UQ9, FE, NDP, ZN, 8Q1, COO, HEM, UQ5, UQ7 |
 |
Cryo-Em Structure Of Nadh:Ubiquinone Oxidoreductase (Complex-I) From Respiratory Supercomplex Of Tetrahymena Thermophila
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: ELECTRON TRANSPORT Ligands: CDL, UDP, MG, PC1, NDP, 3PE, LPP, ADP, FES, SF4, FMN, 8Q1, ZN |
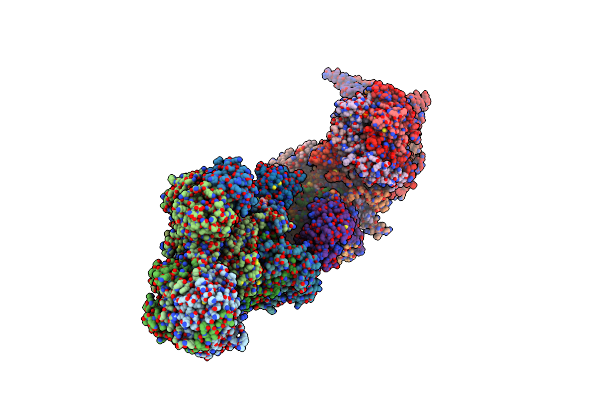 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: ELECTRON TRANSPORT Ligands: SF4, FMN, NAI, PEE, PLX, 8Q1, NDP, UQ, FES, MG, CDL, ZN, ADP |
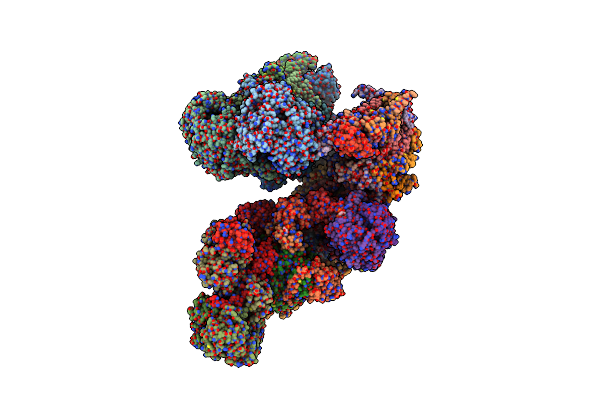 |
Cryo-Em Structure Of The Arabidopsis Thaliana I+Iii2 Supercomplex (Complete Conformation 1 Composition)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-03-22 Classification: MEMBRANE PROTEIN Ligands: SF4, FES, FMN, UQ9, FE, NDP, ZN, 8Q1, COO, HEM, UQ5, UQ7 |
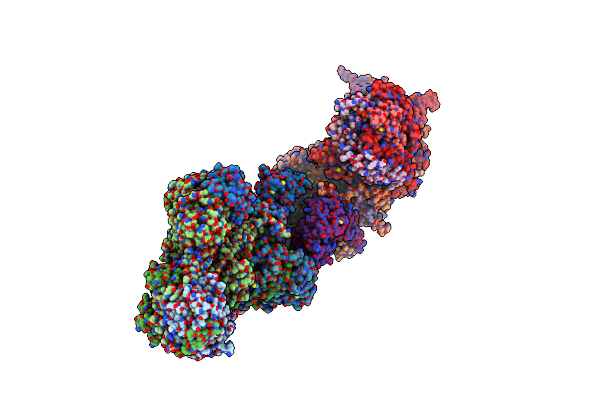 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: ELECTRON TRANSPORT Ligands: SF4, FMN, NAI, PEE, 8Q1, CDL, NDP, FES, MG, ZN, PLX, UQ, ADP |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: ELECTRON TRANSPORT Ligands: SF4, FMN, NAI, PEE, PLX, DCQ, 8Q1, NDP, UQ, FES, MG, CDL, ZN, ADP |


