Search Count: 31
 |
Organism: Erythrobacter sanguineus
Method: ELECTRON MICROSCOPY Resolution:2.27 Å Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: BCL, A1EL2, A1ELD, 8K6, PGV, LMT, BPH, UQ8, FE, H4X, PEF, PO4, LHG |
 |
Organism: Mycolicibacterium
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-10-01 Classification: HYDROLASE Ligands: GOL, MG, 8K6, SO4 |
 |
Cryo-Em Structure Of The Myxol-Bound Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Resolution:2.48 Å Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, A1L4O, CL, PC1, PLC, R16, 8K6, D12, C14, D10 |
 |
Cryo-Em Structure Of The Light-Driven Chloride Ion-Pumping Rhodopsin, Nm-R3
Organism: Nonlabens marinus s1-08
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: RET, CL, PC1, PLC, D12, R16, 8K6, C14 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: EUJ, HEX, D12, D10, OCT, PLC, A1IV3, 8K6 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: OCT, D12, EUJ, A1IV5, 8K6 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: OCT, D12, D10, 8K6, HEX, EUJ, A1IV6 |
 |
Cryo-Em Structure Of Photosynthetic Lh1-Rc Core Complex Of Roseospirillum Parvum
Organism: Roseospirillum parvum
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PHOTOSYNTHESIS Ligands: HEC, MG, PGV, 8K6, BCL, BPH, LMT, FE, MQ8, CRT |
 |
Organism: Thermochromatium tepidum
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: PHOTOSYNTHESIS Ligands: BCL, 8K6, CA, CRT |
 |
 |
Fusidic Acid Binding To The Allosteric Deep Transmembrane Domain Binding Pocket, Tm7/Tm8 Groove, And Tm1/Tm2 Groove Of The Fully Induced Acrb T Protomer
Organism: Escherichia coli (strain k12), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-05-19 Classification: TRANSPORT PROTEIN Ligands: LMT, EDO, GOL, D10, FUA, PGE, 8K6, SO4, PTY, D12, HEX |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-05-19 Classification: TRANSPORT PROTEIN Ligands: LMT, D10, EDO, DDR, MYS, 8K6, MIY, GOL, OCT, SO4, D12, PTY, C14, DDQ |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: ZMA, NA, CLR, D12, MYS, HEX, 8K6, D10, OCT, UND, ER0, TRD |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: ZMA, NA, CLR, D12, MYS, HEX, 8K6, D10, OCT, UND, ER0, TRD |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: ZMA, NA, CLR, D12, MYS, HEX, 8K6, D10, OCT, UND, ER0, TRD |
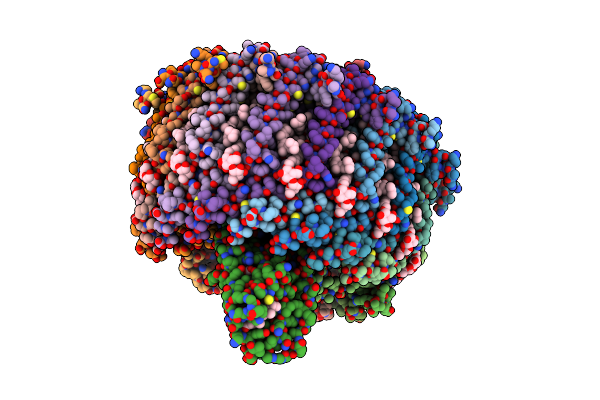 |
Structure Of Photosynthetic Lh1-Rc Super-Complex Of Thiorhodovibrio Strain 970
Organism: Thiorhodovibrio sp. 970
Method: ELECTRON MICROSCOPY Resolution:2.82 Å Release Date: 2020-10-07 Classification: PHOTOSYNTHESIS Ligands: HEM, MG, CA, DGA, PGV, BCL, BPH, UQ8, 8K6, FE, MQ8, CDL, H4X, LMT |
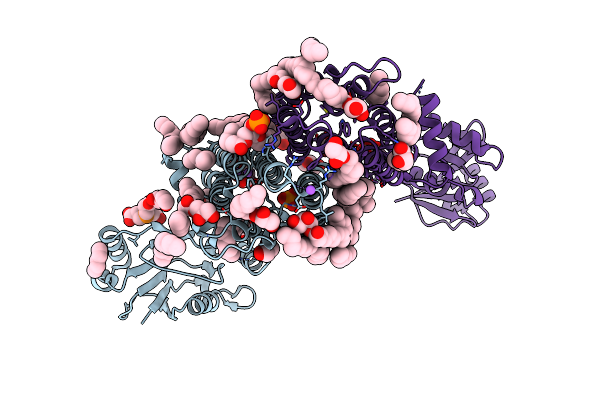 |
Structure Of A Phosphatidylinositol-Phosphate Synthase (Pips) From Mycobacterium Kansasii
Organism: Archaeoglobus fulgidus, Mycobacterium kansasii
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: FLC, NA, PO4, TCE, OLC, 8K6, GOL, XP4 |
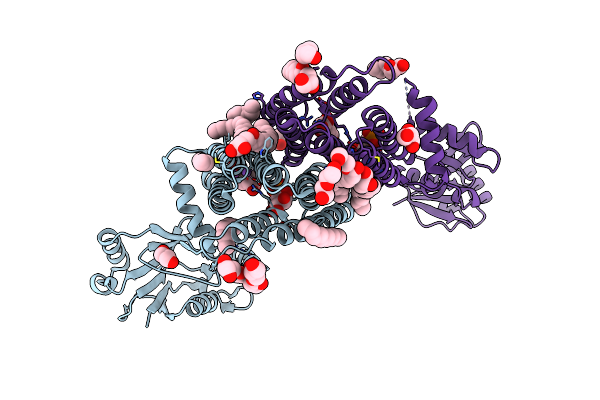 |
Structure Of A Phosphatidylinositol-Phosphate Synthase (Pips) From Mycobacterium Kansasii With Evidence Of Substrate Binding
Organism: Archaeoglobus fulgidus, Mycobacterium kansasii atcc 12478
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2020-05-27 Classification: MEMBRANE PROTEIN Ligands: LIP, NA, TCE, 1PE, OLC, 8K6, GOL, C5P, FLC |
 |
Organism: Sindbis virus
Method: ELECTRON MICROSCOPY Release Date: 2019-03-13 Classification: VIRUS Ligands: 8K6 |
 |
Aftmem16 Reconstituted In Nanodiscs In The Presence Of Ca2+ And Ceramide 24:0
Organism: Neosartorya fumigata (strain atcc mya-4609 / af293 / cbs 101355 / fgsc a1100)
Method: ELECTRON MICROSCOPY Release Date: 2019-02-06 Classification: LIPID TRANSPORT Ligands: CA, D12, D10, 8K6, HEX |

