Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
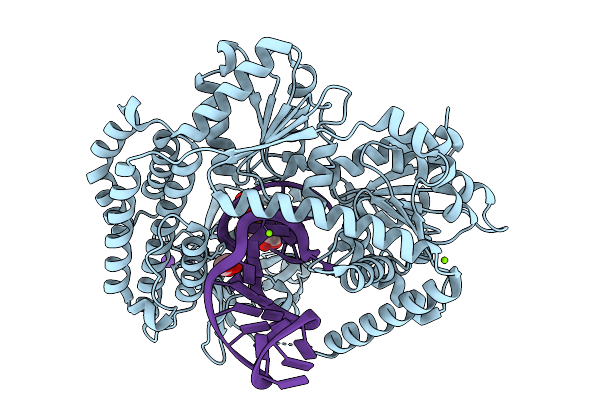 |
Structure Of Duck Rig-I (Delta Cards) Bound To 31-Mer Rna Mismatched Hairpin With 5'Pppg-C Blunt End, Mimicking The Influenza B Virus Vrna Promoter (Panhandle).
Organism: Anas platyrhynchos, Influenza b virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: RNA BINDING PROTEIN Ligands: ZN, MG, CL, K, GOL, GTP |
 |
Structure Of Duck Rig-I (Delta Cards) Bound To 31-Mer Rna Mismatched Hairpin With 5'Ppa-U Blunt End Mimicking The Influenza B Virus Vrna Promoter (Panhandle) And To Adp.
Organism: Anas platyrhynchos, Influenza b virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: RNA BINDING PROTEIN Ligands: ZN, ADP, GOL, K, MG |
 |
Structure Of Duck Rig-I (Delta Cards) Bound To 31-Mer Rna Mismatched Hairpin With 5'Pppg-C Blunt End Mimicking The Influenza A Virus Vrna Promoter (Panhandle) And To Adp.
Organism: Anas platyrhynchos, Influenza a virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: RNA BINDING PROTEIN Ligands: ZN, ADP, CL, MG, GTP |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN |
 |
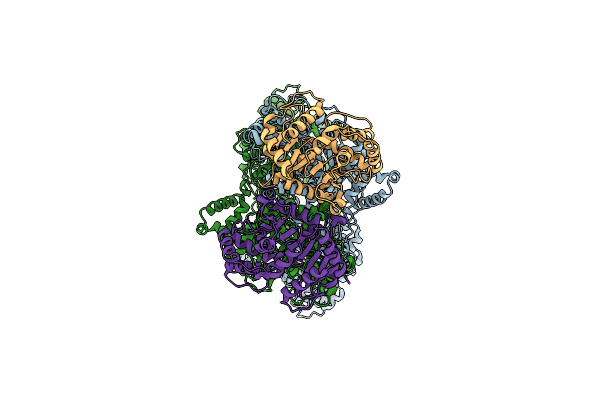
Organism: Streptomyces
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN |
 |
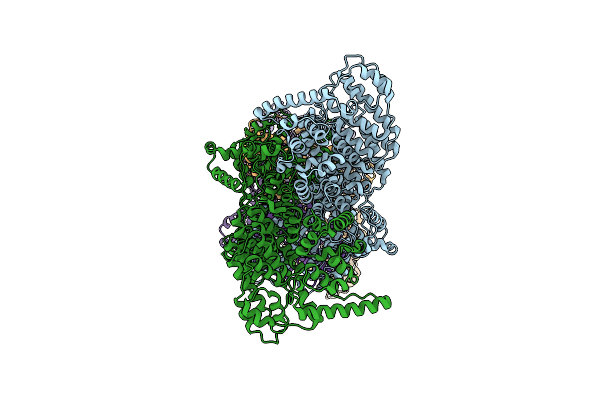
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |