Search Count: 1,763
 |
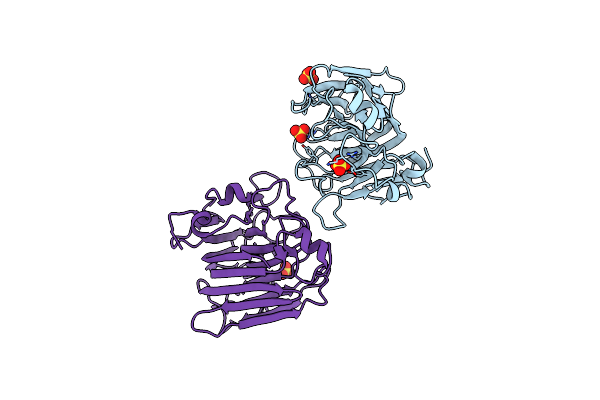
Organism: Pseudomonas pavonaceae, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: LIGASE Ligands: G2P, MG, TA1 |
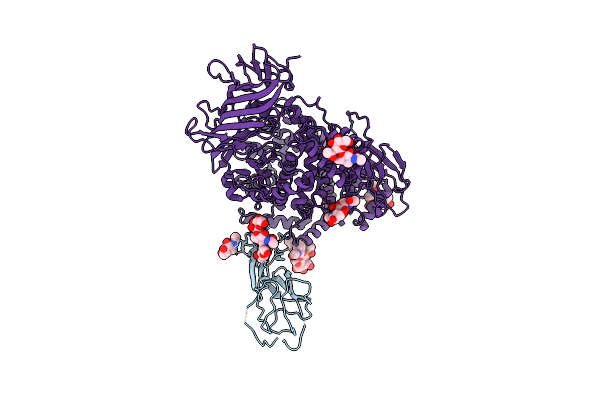 |
Organism: Tgev virulent purdue, Canis lupus familiaris
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: ISOMERASE Ligands: NAG |
 |
Organism: Saccharina japonica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-16 Classification: LYASE Ligands: SO4 |
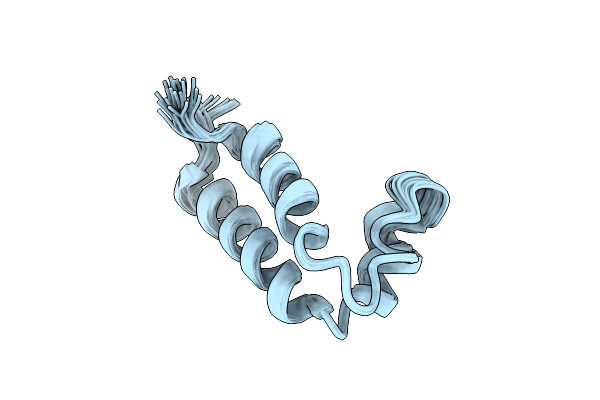 |
Organism: Apilactobacillus kunkeei
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: UNKNOWN FUNCTION |
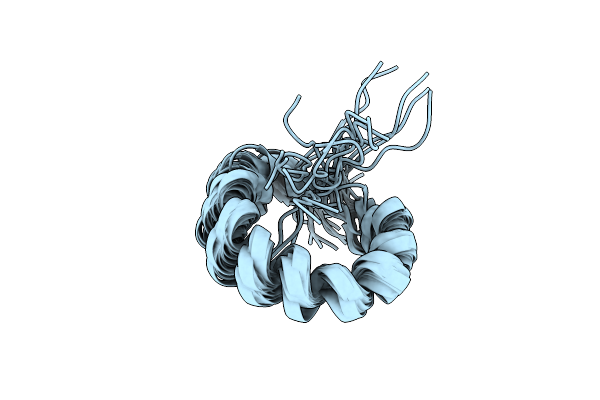 |
Organism: Apilactobacillus kunkeei
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: UNKNOWN FUNCTION |
 |
Organism: Apilactobacillus kunkeei
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: UNKNOWN FUNCTION |
 |
Organism: Saccharum hybrid cultivar
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-10-18 Classification: PLANT PROTEIN Ligands: ATP, MG |
 |
Organism: Saccharum hybrid cultivar
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2023-10-18 Classification: PLANT PROTEIN Ligands: ATP, HX2, FMT |
 |
Organism: Saccharum hybrid cultivar
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-10-18 Classification: PLANT PROTEIN Ligands: ADP, GOL |
 |
Organism: Desulfovibrio vulgaris str. hildenborough, Desulfovibrio vulgaris
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: DNA BINDING PROTEIN/DNA/RNA |
 |
Structural, Kinetic, And Mechanistic Analysis Of The Wild-Type And Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis For The Decarboxylase And Hydratase Activities
Organism: Pseudomonas pavonaceae
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-12-21 Classification: ISOMERASE |
 |
Organism: Hiv-1 o_mvp5180
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-10-19 Classification: VIRAL PROTEIN |
 |
Xenon-Bound Structure Of Carbon Monoxide Dehydrogenase (Codh) From Desulfovibrio Vulgaris
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-05-04 Classification: OXIDOREDUCTASE Ligands: SF4, FES, CUV, XE |
 |
Organism: Parvibaculum lavamentivorans ds-1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-02-02 Classification: OXIDOREDUCTASE |
 |
Oxidized Thioredoxin 1 From The Anaerobic Bacteria Desulfovibrio Vulgaris Hildenborough
Organism: Desulfovibrio vulgaris
Method: SOLUTION NMR Release Date: 2021-07-14 Classification: OXIDOREDUCTASE |
 |
Organism: Influenza a virus (a/hong kong/483/1997(h5n1)), Unidentified influenza virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-04-14 Classification: VIRAL PROTEIN/RNA Ligands: EDO |
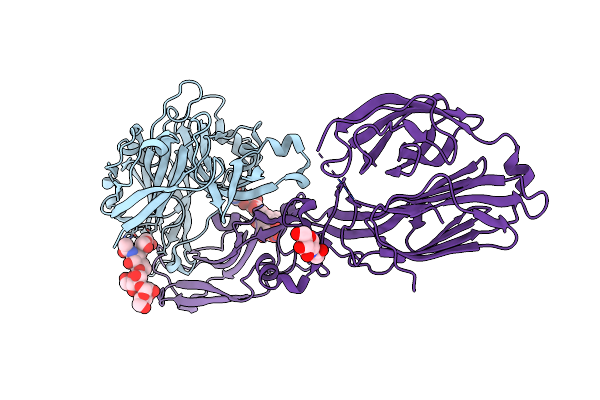 |
Crystal Structure Of The Envelope Glycoprotein Complex Of Maporal Virus In A Prefusion Conformation
Organism: Maporal virus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-10-14 Classification: VIRAL PROTEIN Ligands: NAG |
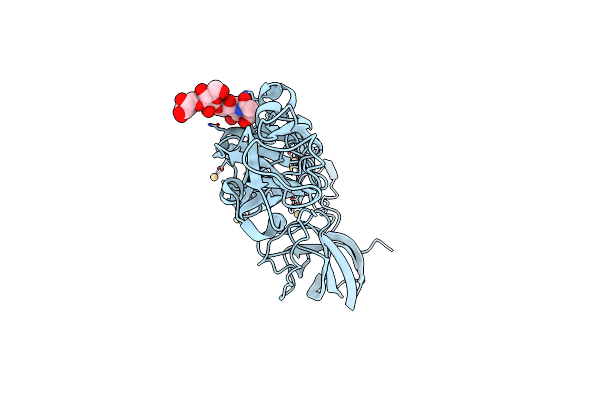 |
Structure Of Maporal Virus Envelope Glycoprotein Gc In Postfusion Conformation
Organism: Maporal virus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-10-14 Classification: VIRAL PROTEIN Ligands: CD |
 |
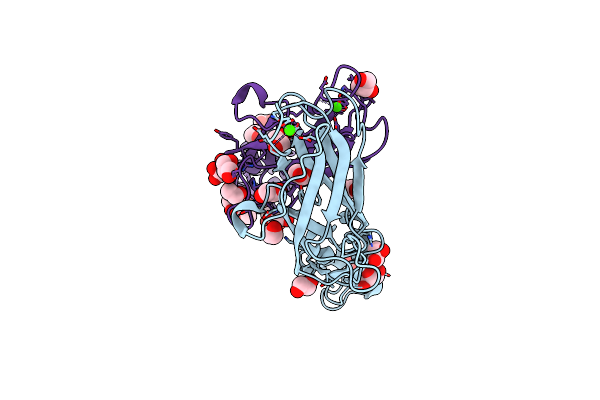
Organism: Alicyclobacillus tengchongensis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-07-29 Classification: HYDROLASE Ligands: B3P, EDO, NA, IOD |
 |
Organism: Streptococcus sanguinis sk678
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2020-02-19 Classification: SUGAR BINDING PROTEIN Ligands: CA, CO, GOL, EDO |

