Search Count: 769
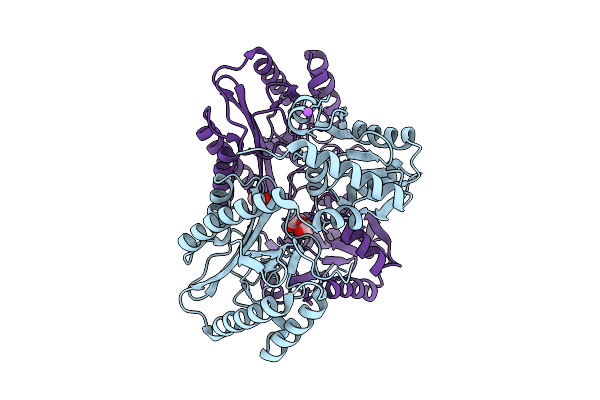 |
Crystal Structure Of Proline Dipeptidase From Pyrococcus Furiosus Complexed With Co
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE |
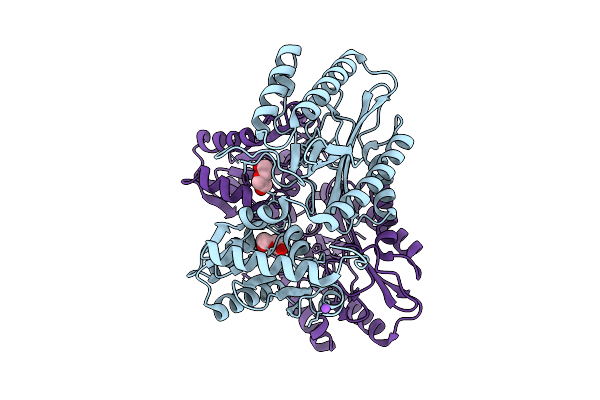 |
Crystal Structure Of Proline Dipeptidase From Pyrococcus Furiosus Complexed With Mn
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MN, NA, PEG |
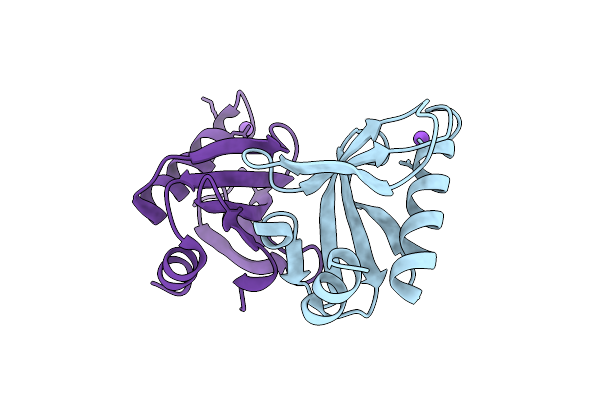 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Sodium Acetate At Ph 8
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
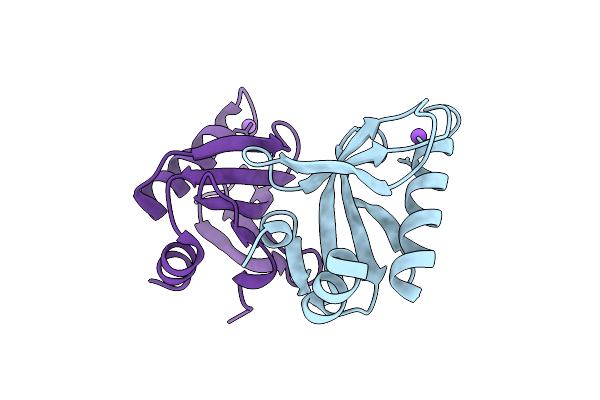 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus At Ph 6
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus At Ph 9
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Sodium Citrate At Ph 8.0
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Peg1500 And Glycerol At Ph 8.0
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION Ligands: GOL, CL |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus At Ph 4
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Cacl2 At Ph 5
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Cacl2 At Ph 6
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION Ligands: CL, NA |
 |
Crystal Structure Of Upf0235 Protein Pf1765 From Pyrococcus Furiosus Crystallized With Cacl2 At Ph 7
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN |
 |
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: UMP |
 |
Dolichyl Phosphate Mannose Synthase In Complex With Donor (Gdp-Man) And Traces Of Acceptor (Dol55P) And Product (Dol55P-Man)
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: GDD, MG, EFS, MJC |
 |
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: UNKNOWN FUNCTION |
 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Naja kaouthia, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TOXIN Ligands: CL, GOL, SO4, NA |
 |
Structure Of The Keops Complex (Cgi121/Bud32/Kae1/Pcc1) Bound To Trna In Its Native-Like Conformation.
Organism: Methanocaldococcus jannaschii, Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: RNA BINDING PROTEIN/RNA |
 |
Structure Of The Keops Complex (Cgi121/Bud32/Kae1/Pcc1) Bound To Trna In A Distorted Trna Conformation
Organism: Methanocaldococcus jannaschii, Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Synthetic construct, Naja kaouthia
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2024-10-09 Classification: DE NOVO PROTEIN/TOXIN |

