Search Count: 607
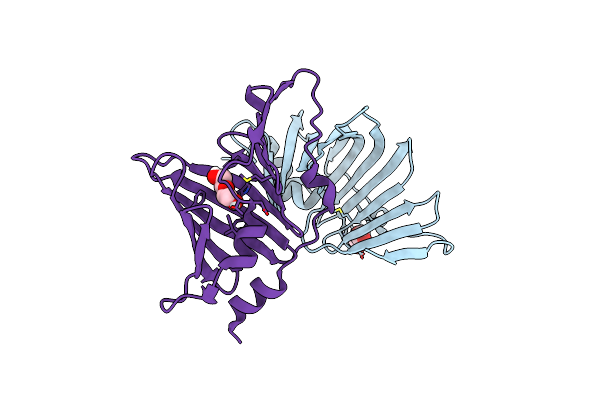 |
Structural And Functional Analysis Of The Helicobacter Pylori Lipoprotein Chaperone Lola
Organism: Helicobacter pylori j99
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-12-18 Classification: TRANSPORT PROTEIN Ligands: PEG |
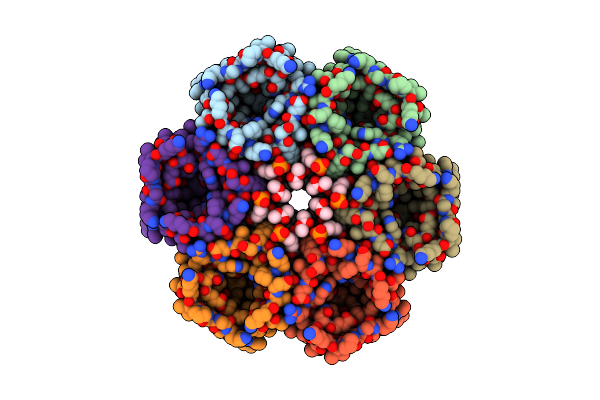 |
Organism: Helicobacter pylori (strain j99 / atcc 700824)
Method: ELECTRON MICROSCOPY Release Date: 2019-04-03 Classification: TRANSPORT PROTEIN Ligands: XP4 |
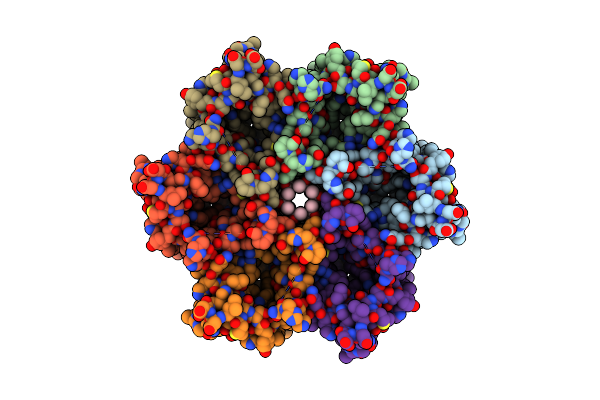 |
Organism: Helicobacter pylori (strain j99 / atcc 700824)
Method: ELECTRON MICROSCOPY Release Date: 2019-04-03 Classification: TRANSPORT PROTEIN Ligands: XP4 |
 |
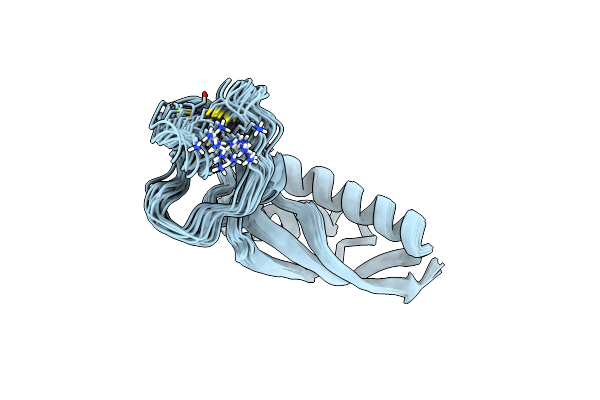
N-Terminal Domain Of Translation Initiation Factor If-3 From Helicobacter Pylori
Organism: Helicobacter pylori j99
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2019-03-13 Classification: LIGASE |
 |
Solution Structure Of The Ni Metallochaperone Hypa From Helicobacter Pylori
Organism: Helicobacter pylori j99
Method: SOLUTION NMR Release Date: 2018-10-10 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2018-06-27 Classification: DNA BINDING PROTEIN |
 |
Organism: Helicobacter pylori (strain j99 / atcc 700824)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2017-12-06 Classification: UNKNOWN FUNCTION Ligands: EPE, CA |
 |
Organism: Helicobacter pylori (strain j99 / atcc 700824)
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2016-11-23 Classification: HYDROLASE Ligands: SAH, DOD |
 |
1.4 A Resolution Structure Of Helicobacter Pylori Mtan In Complexed With P-Clph-Dadme-Imma
Organism: Helicobacter pylori (strain j99 / atcc 700824)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-11-16 Classification: HYDROLASE Ligands: 4CT, MG |
 |
Crystal Structures Of Ferritin Mutants Reveal Side-On Binding To Diiron And End-On Cleavage Of Oxygen
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-07-27 Classification: OXIDOREDUCTASE Ligands: FE, IMD |
 |
Crystal Structure Of Helicobacter Pylori 5'-Methylthioadenosine/S-Adenosyl Homocysteine Nucleosidase (Mtan) Complexed With Methylthio-Dadme-Immucillin-A
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-11-25 Classification: hydrolase/hydrolase inhibitor Ligands: TDI |
 |
Crystal Structure Of Helicobacter Pylori 5'-Methylthioadenosine/S-Adenosyl Homocysteine Nucleosidase (Mtan) Complexed With Hydroxybutylthio-Dadme-Immucillin-A
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-11-25 Classification: hydrolase/hydrolase inhibitor Ligands: GMD |
 |
Crystal Structure Of Helicobacter Pylori 5'-Methylthioadenosine/S-Adenosyl Homocysteine Nucleosidase (Mtan) Complexed With 2-(2-Hydroxyethoxy)Ethylthiomethyl-Dadme-Immucillin-A
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2015-11-25 Classification: hydrolase/hydrolase inhibitor Ligands: 3QA, SO4 |
 |
Crystal Structure Of Helicobacter Pylori 5'-Methylthioadenosine/S-Adenosyl Homocysteine Nucleosidase (Mtan) Complexed With Pyrazinylthio-Dadme-Immucillin-A
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-11-25 Classification: Hydrolase/inhibitor Ligands: 4EH, GOL, PEG |
 |
Crystal Structure Of Helicobacter Pylori 5'-Methylthioadenosine/S-Adenosyl Homocysteine Nucleosidase (Mtan) Complexed With (((4-Amino-5H-Pyrrolo[3,2-D]Pyrimidin-7-Yl)Methyl)(Hexyl)Amino)Methanol
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-11-25 Classification: Hydrolase/Hydrolase Inhibitor Ligands: 4EZ, ZN |
 |
Crystal Structure Of Molybdenum Cofactor Biosynthesis Protein Moga From Helicobacter Pylori Str. J99
Organism: Helicobacter pylori (strain j99 / atcc 700824)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-02-04 Classification: TRANSFERASE |
 |
Crystal Structure Of The Helicobacter Pylori Mtan-D198N Mutant With S-Adenosylhomocysteine In The Active Site
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2014-12-17 Classification: HYDROLASE Ligands: SAH, CL |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-09-10 Classification: HYDROLASE |
 |
1.88 Angstrom Resolution Crystal Structure Of Hypothetical Protein Jhp0584 From Helicobacter Pylori.
Organism: Helicobacter pylori j99
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2014-09-10 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2014-04-09 Classification: TRANSFERASE |

