Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
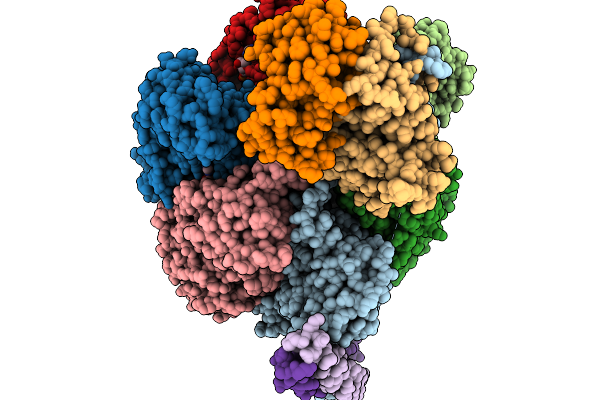 |
Cryo-Em Structure Of F-Atp Synthase From Mycobacteroides Abscessus (Rotational State 1)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ATP, MG, ADP |
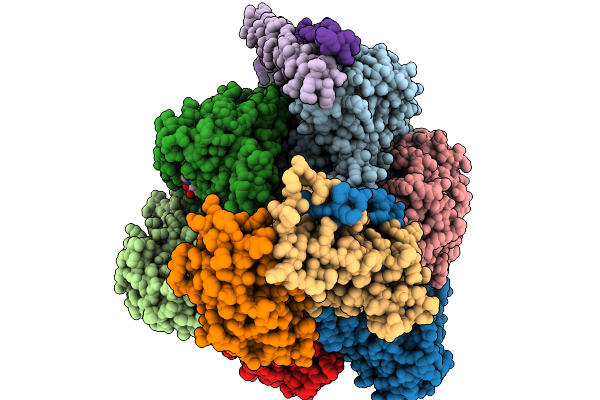 |
Cryo-Em Structure Of F-Atp Synthase From Mycobacteroides Abscessus (Rotational State 2)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ATP, MG, ADP |
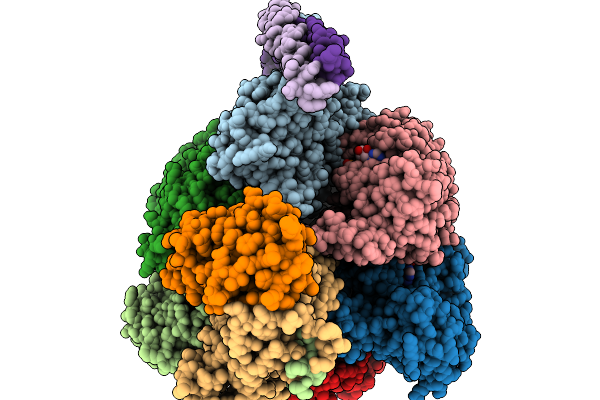 |
Cryo-Em Structure Of F-Atp Synthase From Mycobacteroides Abscessus (Rotational State 3)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ATP, MG, ADP |
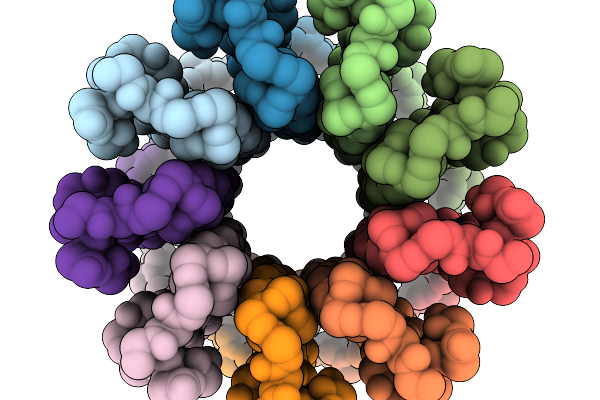 |
Cryo-Em Structure Of F-Atp Synthase C-Ring From Mycobacteroides Abscessus (Backbone)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: HYDROLASE |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SIGNALING PROTEIN Ligands: CL, ETA |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SIGNALING PROTEIN Ligands: 2A1, CL |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SIGNALING PROTEIN Ligands: SEL, CL |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: SIGNALING PROTEIN |
 |
3Dflex Refinement Of The Cryoem Structure Of Declic Nanodisc With 10Mm Edta In Sym-Like State
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TRANSLOCASE |
 |
3Dflex Refinement Of The Cryoem Structure Of Declic Nanodisc With 10Mm Edta In Asym State
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TRANSLOCASE |
 |
Non-Uniform Refinement Of The Cryoem Structure Of Declic Nanodisc With 10Mm Calcium
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TRANSLOCASE Ligands: CA, OCT, D12, D10, C14, PX6 |
 |
Non-Uniform Refinement Of The Cryoem Structure Of Declic Nanodisc With 10Mm Edta In Asym State
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TRANSLOCASE Ligands: OCT, D10, C14 |
 |
3Dflex Refinement Of The Cryoem Structure Of Declic Nanodisc With 10Mm Calcium
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TRANSLOCASE Ligands: CA |
 |
Non-Uniform Refinement Of The Cryoem Structure Of Declic Nanodisc With 10Mm Edta In Sym-Like State
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TRANSLOCASE Ligands: OCT, D10, C14 |
 |
Organism: Human herpesvirus 8 strain gk18
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: VIRAL PROTEIN |
 |
Closed Conformation Of The Pentameric Ligand-Gated Ion Channel, Declic At Ph 5 With 10 Mm Ca2+
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
Open Conformation Of The Pentameric Ligand-Gated Ion Channel, Declic At Ph 5 With 10Mm Ca2+
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN |
 |
Open Conformation Of The Pentameric Ligand-Gated Ion Channel, Declic At Ph 5 With 10Mm Edta
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN |
 |
Closed And Disordered Conformation Of The Pentameric Ligand-Gated Ion Channel, Declic At Ph 5 With 10Mm Edta
Organism: Desulfofustis sp. pb-srb1
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN |
 |
Organism: Acinetobacter sp. yt-02
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-09-04 Classification: FLAVOPROTEIN Ligands: FAD |