Search Count: 28
 |
Organism: Moesziomyces antarcticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: CL, LYS |
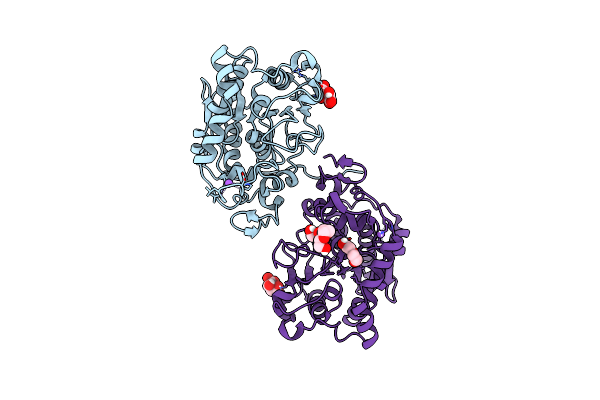 |
Crystal Structure Of Candida Antarctica Lipase B With The Putative Pro-Peptide Region
Organism: Moesziomyces antarcticus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-04-09 Classification: LYASE Ligands: TLA, NA, PEG, PG4 |
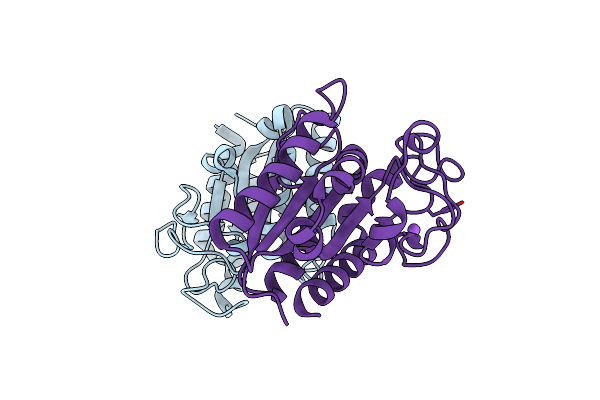 |
A Biodegradable Plastic-Degrading Cutinase-Like Enzyme From The Phyllosphere Yeast Pseudozyma Antarctica
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-15 Classification: HYDROLASE |
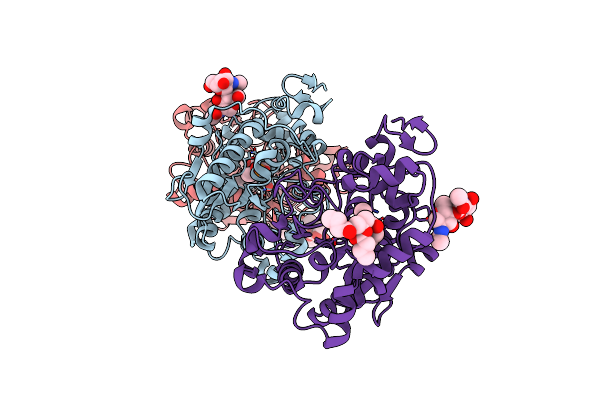 |
Substrate Protein Interactions In The Limbus Region Of The Catalytic Site Of Candida Antarctica Lipase B
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: DEP, NTK |
 |
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-01-01 Classification: HYDROLASE Ligands: SO4, EDO, ACT, CL, PGE |
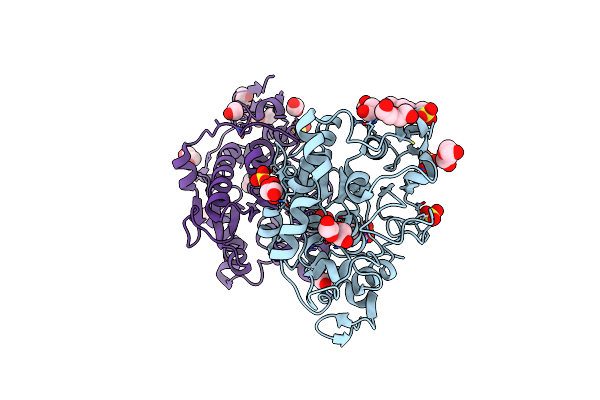 |
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-01-01 Classification: HYDROLASE Ligands: SO4, EDO, EPE, PEG, CL, 1PE |
 |
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-01-01 Classification: HYDROLASE Ligands: SO4, EDO, PEG |
 |
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2020-01-01 Classification: HYDROLASE Ligands: SO4, PEG, EDO |
 |
Crystal Structure Of Candida Antarctica Lipase B Mutant Sr With Product Analogue
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2020-01-01 Classification: HYDROLASE Ligands: B7U, SO4, PGE, EDO, CL, PEG |
 |
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: CPQ, CA |
 |
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: ACT, IPA, EDO, PGE |
 |
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: NI, PEG, PG4 |
 |
Crystal Structure Of Candida Antarctica Lipase B With Active Ser105 Modified With A Phosphonate Inhibitor
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2017-09-13 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: MSW |
 |
Crystal Structure Of S. Pombe Ubiquitin E1 (Uba1) In Complex With Ubc15 And Ubiquitin
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843), Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-02-15 Classification: LIGASE Ligands: SO4 |
 |
Open And Closed Conformations And Protonation States Of Candida Antarctica Lipase B: Xenon Complex
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2015-10-21 Classification: HYDROLASE Ligands: IPA, XE |
 |
Open And Closed Conformations And Protonation States Of Candida Antarctica Lipase B: Atomic Resolution Native
Organism: Pseudozyma antarctica
Method: X-RAY DIFFRACTION Resolution:0.91 Å Release Date: 2015-10-21 Classification: HYDROLASE Ligands: IPA, K, NA |
 |
 |
 |
Organism: Eupithecia sp. BOLD:AAB1831
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Eupithecia sp. BOLD:ACM4052
Method: Alphafold Release Date: Classification: NA Ligands: NA |

