Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Phocaeicola plebeius dsm 17135
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-10 Classification: HYDROLASE Ligands: GOL, CA, CL, ACT |
 |
Organism: Phocaeicola plebeius dsm 17135
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: EPE, ACT, CA, PEG |
 |
Organism: Phocaeicola plebeius dsm 17135
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: EDO, P4G, PG4, PEG, CIT, NA |
 |
Organism: Phocaeicola plebeius dsm 17135
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: PO4, K |
 |
Organism: Phocaeicola plebeius dsm 17135
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-10-05 Classification: HYDROLASE Ligands: PEG, EDO, PG4, PGE, NA |
 |
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-02-09 Classification: OXIDOREDUCTASE |
 |
Structure Of The Exo-Alpha-L-Galactosidase Bpgh29 From Bacteroides Plebeius
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: EDO, CA |
 |
Structure Of The Exo-Alpha-L-Galactosidase Bpgh29 From Bacteroides Plebeius In Complex With L-Galactose
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, GXL, EDO |
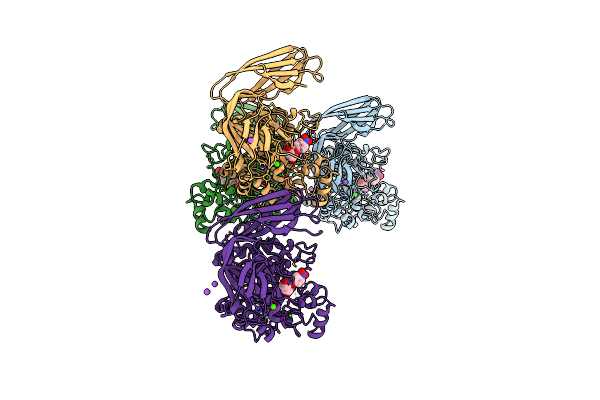 |
Structure Of The Exo-Alpha-L-Galactosidase Bpgh29 (D264N Mutant) From Bacteroides Plebeius In Complex With Paranitrophenyl-Alpha-L-Galactopyranoside
Organism: Bacteroides plebeius
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-02-09 Classification: HYDROLASE Ligands: CA, Y9Y, NI, NA, CL |
 |
Structure Of Thioredoxin Reductase From The Thermophilic Eubacterium Thermosipho Africanus Tcf52B
Organism: Thermosipho africanus (strain tcf52b)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-10-06 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
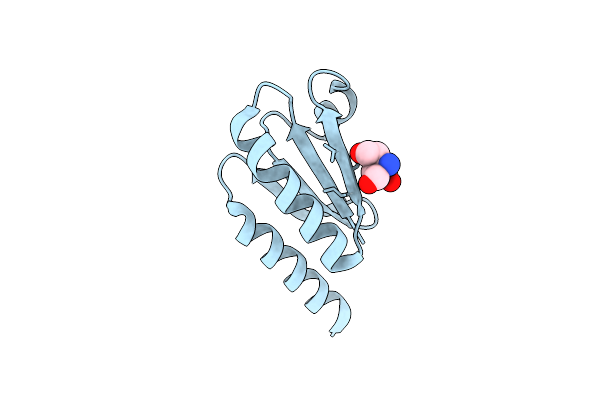 |
Structure Of Thioredoxin 1 From The Thermophilic Eubacterium Thermosipho Africanus Tcf52B
Organism: Thermosipho africanus (strain tcf52b)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-08-14 Classification: OXIDOREDUCTASE Ligands: TAM |
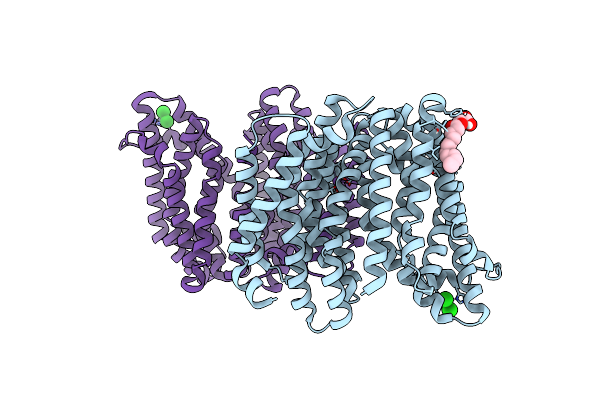 |
Organism: Thermosipho africanus (strain tcf52b)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-04-17 Classification: TRANSPORT PROTEIN |
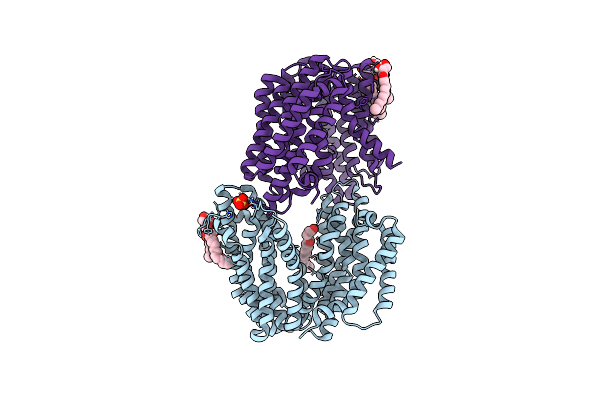 |
Organism: Thermosipho africanus (strain tcf52b)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-04-17 Classification: TRANSPORT PROTEIN Ligands: OLC, SO4 |
 |
Organism: Thermosipho africanus (strain tcf52b)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-04-17 Classification: TRANSPORT PROTEIN |
 |
Organism: Thermosipho africanus (strain tcf52b)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-04-17 Classification: TRANSPORT PROTEIN Ligands: 1PE, OLB, NA, OLC |
 |
Organism: Orpinomyces sp. y102
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: GOL, PO4, EDO, MPD |
 |
Organism: Orpinomyces sp. y102
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-05-17 Classification: HYDROLASE Ligands: PO4, GOL, EDO, NH4, MPD, PEG, ACT |
 |
Organism: Thermosipho africanus (strain tcf52b)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-12-28 Classification: TRANSPORT PROTEIN Ligands: CL, ZN, CA, NA, 1PE, OLB, OLC |
 |
 |