Search Count: 4,586
 |
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: TRANSPORT PROTEIN |
 |
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: TRANSPORT PROTEIN |
 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN |
 |
Organism: Xenopus laevis, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: GENE REGULATION |
 |
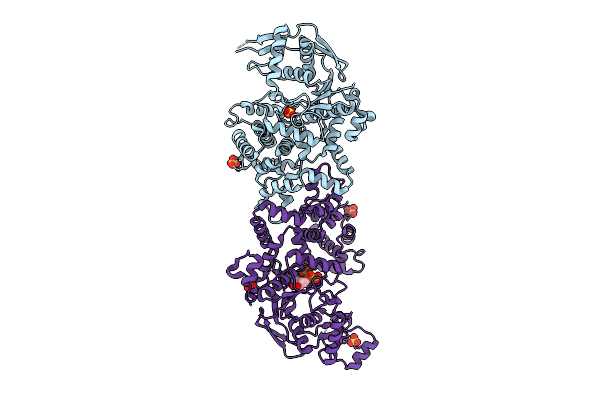
Crystal Structure Of Glutamate-Trna Synthetase Glurs From Chlamydia Pneumoniae (Orthorhombic C Form)
Organism: Chlamydia pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LIGASE Ligands: CL |
 |
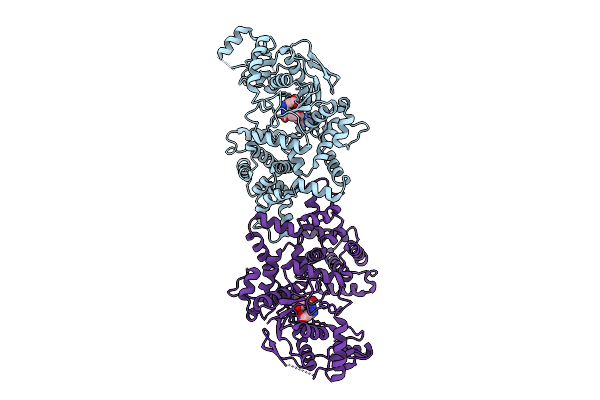
Crystal Structure Of Glutamate-Trna Synthetase Glurs From Chlamydia Pneumoniae In Complex With Atp
Organism: Chlamydia pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LIGASE Ligands: SO4, CL, ATP |
 |
Crystal Structure Of Glutamate-Trna Synthetase Glurs From Chlamydia Pneumoniae In Complex With O5'-(L-Glutamyl-Sulfamoyl)-Adenosine
Organism: Chlamydia pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LIGASE Ligands: GSU, CL |
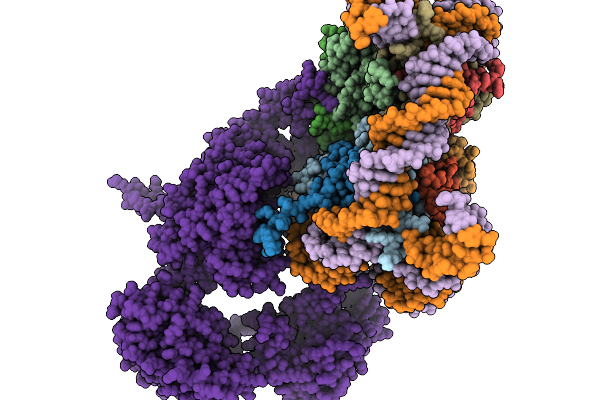 |
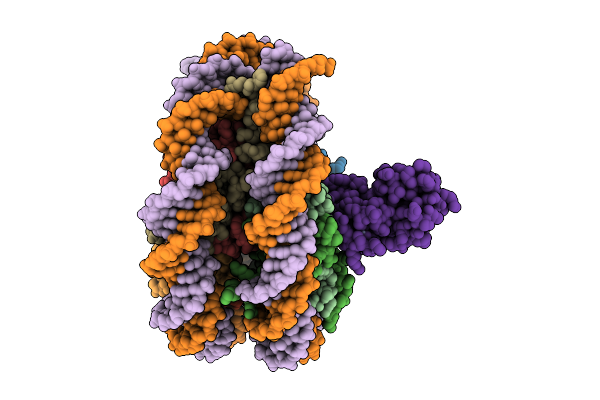
Organism: Xenopus laevis, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DNA BINDING PROTEIN |
 |
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Symmetry-Expanded Reconstruction Of Augmin T-Ii Bonsai On The Gtpgammas Microtubule
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CELL CYCLE Ligands: GTP, MG, GSP |
 |
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CELL CYCLE Ligands: G2P, MG, GTP |
 |
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN |
 |
Structure Of Xenopus Kcnq1(E150R/R221E)-Cam With The Vsd In The Intermediate State
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN |
 |
The Cryo-Em Structure Of Nucleosome-Bound Dna Methyltransferases Dnmt3A2 And Dnmt3L
Organism: Xenopus laevis, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: DNA BINDING PROTEIN Ligands: ZN, SAH |
 |
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Synthetic construct, Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Xenopus laevis, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Chromosomal Passenger Complex In Complex With H3T3Ph Nucleosome (Double Occupancy)
Organism: Homo sapiens, Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CELL CYCLE |
 |
Organism: Xenopus laevis, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: NUCLEAR PROTEIN |

