Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
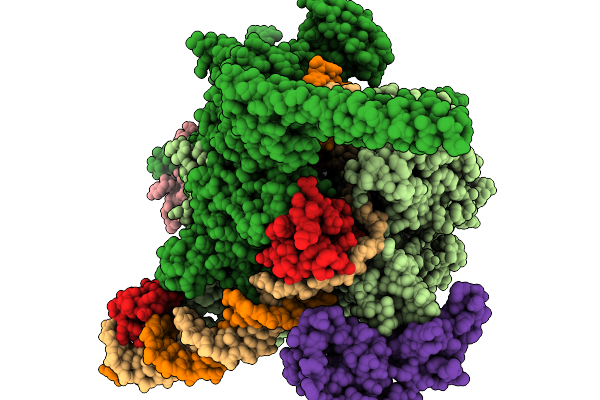 |
Structural Basis Of Pausing During Transcription Initiation In Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: TRANSCRIPTION/DNA/RNA Ligands: MG, ZN |
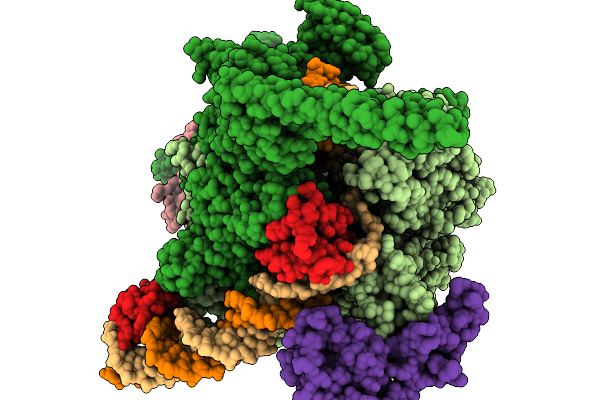 |
Structural Basis Of Pausing During Transcription Initiation In Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: TRANSCRIPTION/DNA/RNA Ligands: MG, ZN |
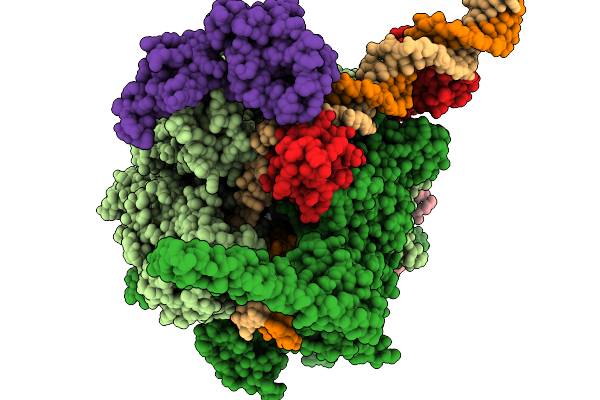 |
Structural Basis Of Pausing During Transcription Initiation In Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: TRANSCRIPTION/DNA/RNA Ligands: MG, ZN |
 |
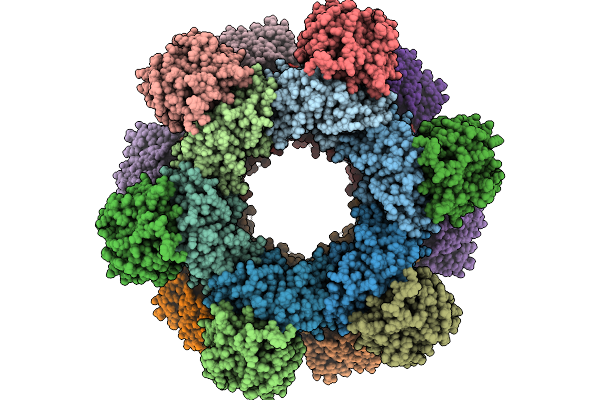
Structures And Mechanism Of The Nicotinamide Nucleotide Transhydrogenase From E. Coli
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: PC1, PKZ |
 |
Cryo-Em Structure Of A Designed Pyridoxal Phosphate (Plp) Synthase Fused To A Designed Circumsporozoite Protein Antigen From Plasmodium Falciparum (Csp-P1-Csp And Csp-P2-Csp)
Organism: Plasmodium falciparum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: BIOSYNTHETIC PROTEIN |
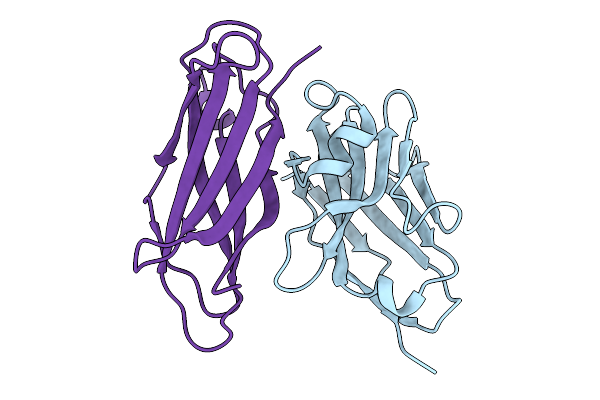 |
Crystal Structure Of Malaria Transmission-Blocking Antigen Pfhap2 Domain 3 In Complex With Nanobody Wnb 334
Organism: Vicugna pacos, Plasmodium falciparum
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: PROTEIN BINDING |
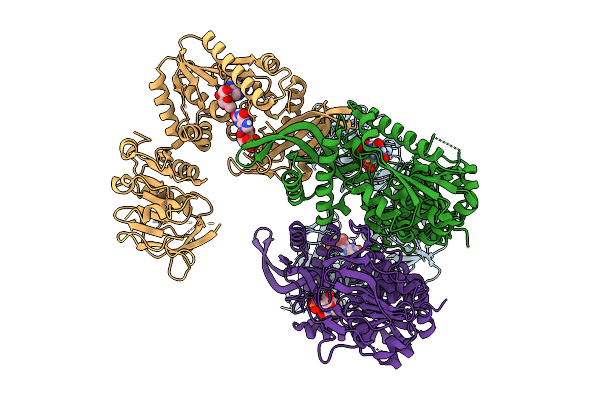 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LIGASE/ANTIBIOTIC Ligands: XMP, A1L67 |
 |
Mycobacterium Tuberculosis Relbe1 Toxin-Antitoxin System; Rv1247C (Relb1 Antitoxin), Rv1246C (Rele1 Toxin)
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TOXIN Ligands: CL |
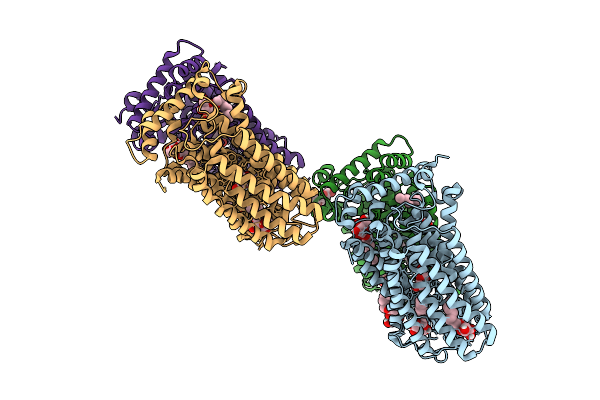 |
Structure Of The Malarial Hexose Transporter 1 (Pfht1) In Complex With 2,5-Anhydro-D-Mannitol.
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: BNG, A1IVP |
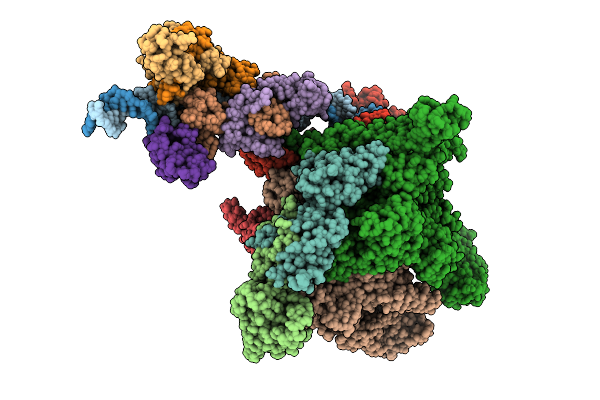 |
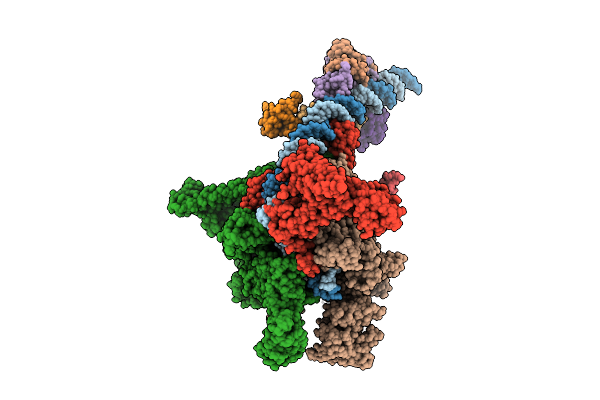
Cryo-Em Structure Of E.Coli Transcription Initiation Complex With Escherichia Phage Mu Late Transcription Activator C
Organism: Escherichia coli (strain k12), Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION/DNA |
 |
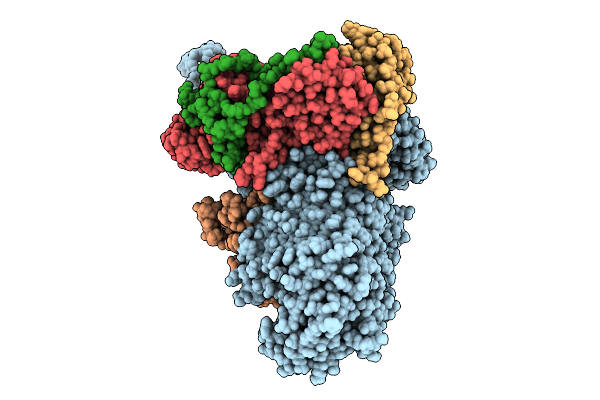
Cryo-Em Structure Of E.Coli Transcription Initiation Complex With Escherichia Phage Mu Middle Transcription Activator Mor
Organism: Escherichia coli (strain k12), Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION/DNA |
 |
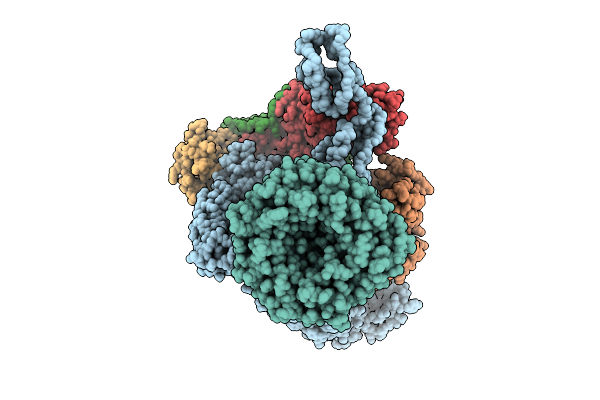
Beta-Barrel Assembly Machine From Escherichia Coli In An Early State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
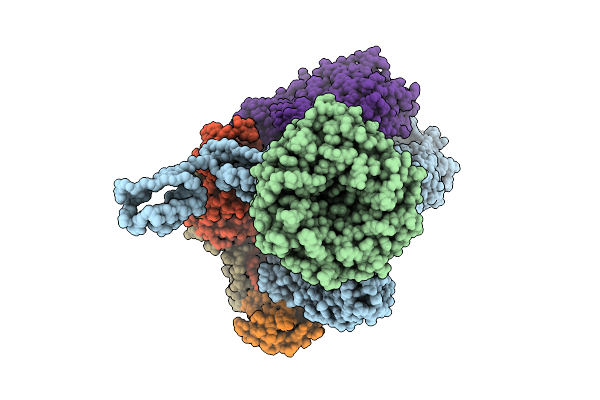
Beta-Barrel Assembly Machine From Escherichia Coli In A Middle State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Beta-Barrel Assembly Machine From Escherichia Coli In A Late State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
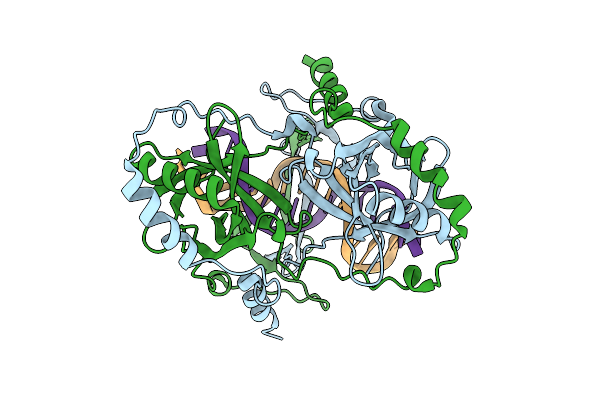 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
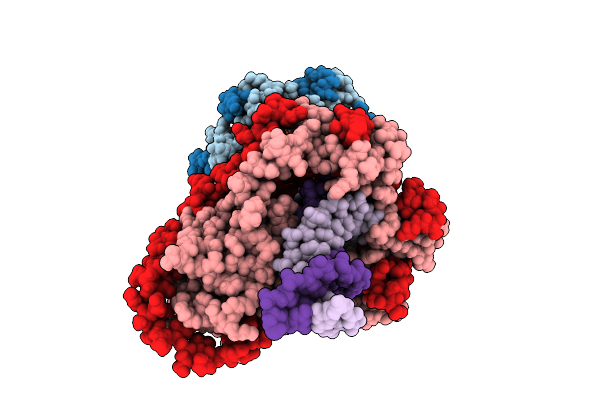 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA |
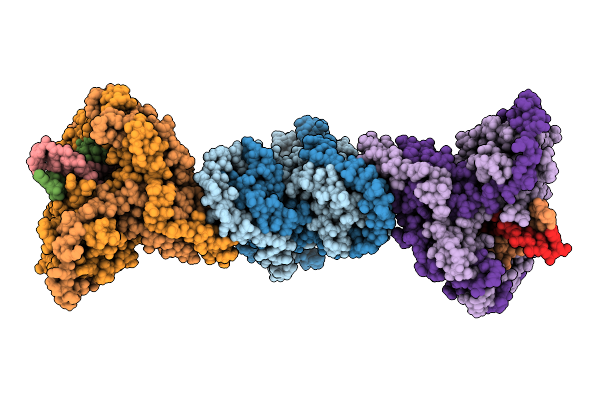 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
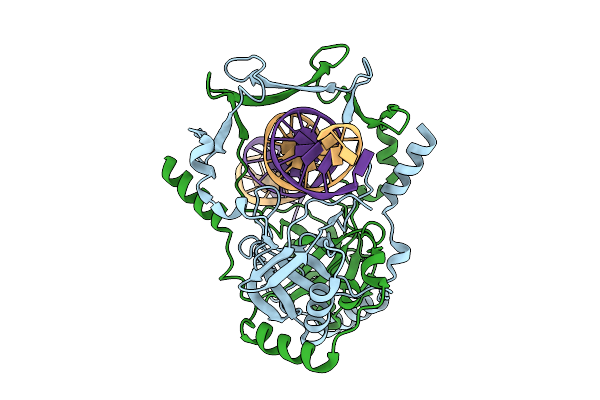 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: ATP, PEG, KAA |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: FE2, FES, HCI |