Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria)
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LIGASE Ligands: CL, A1JC1 |
 |
Organism: Coronavirus neoromicia/pml-phe1/rsa/2011, Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: VIRAL PROTEIN Ligands: NAG, EIC |
 |
Crystal Structure Of Gh29 Family Alpha-L-Fucosidase From Fusarium Proliferatum Le1
Organism: Gibberella intermedia
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: NAG, TRS, MPD, NA, AZI, ACT |
 |
Organism: Tick-borne encephalitis virus-european subtype, Langat virus (strain tp21)
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: VIRUS Ligands: CPL |
 |
Organism: Tick-borne encephalitis virus-european subtype, Langat virus (strain tp21)
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: VIRUS Ligands: CPL |
 |
X-Ray Crystallographic Structure Of Snoal2 In Complex With The Polyketide Substrate
Organism: Streptomyces nogalater
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-08-07 Classification: BIOSYNTHETIC PROTEIN Ligands: XN0 |
 |
X-Ray Crystallographic Structure Of Snoal2 In Complex With The Polyketide Reaction Product
Organism: Streptomyces nogalater
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-08-07 Classification: BIOSYNTHETIC PROTEIN Ligands: XN8 |
 |
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-05 Classification: HYDROLASE Ligands: XSR, CL |
 |
Structure Of The Alginate Epimerase/Lyase Complexed With Penta-Mannuronic Acid
Organism: Azotobacter chroococcum ncimb 8003
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-05-15 Classification: ISOMERASE Ligands: CA, PEG, PGE |
 |
Structure Of The Alginate Epimerase/Lyase Complexed With Tetra-Mannuronic Acid
Organism: Azotobacter chroococcum ncimb 8003
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-05-15 Classification: ISOMERASE Ligands: CA, PEG, ACT, PG4 |
 |
Organism: Azotobacter chroococcum ncimb 8003
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2024-05-08 Classification: ISOMERASE Ligands: CA, PEG, PGE |
 |
Structure Of The Alginate Epimerase/Lyase Complexed With Tri-Mannuronic Acid
Organism: Azotobacter chroococcum ncimb 8003
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-05-08 Classification: ISOMERASE Ligands: CA, ACT |
 |
Structure Of The Alginate Epimerase/Lyase Complexed With Di-Mannuronic Acid
Organism: Azotobacter chroococcum ncimb 8003
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-05-08 Classification: ISOMERASE Ligands: CA |
 |
Cryo-Em Structure Of A Bacterial Nitrilase Filament With A Covalent Adduct Derived From Benzonitrile Hydrolysis
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: HYDROLASE Ligands: HBX |
 |
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria)
Method: X-RAY DIFFRACTION Release Date: 2024-04-17 Classification: HYDROLASE Ligands: TN9, CL, SO4 |
 |
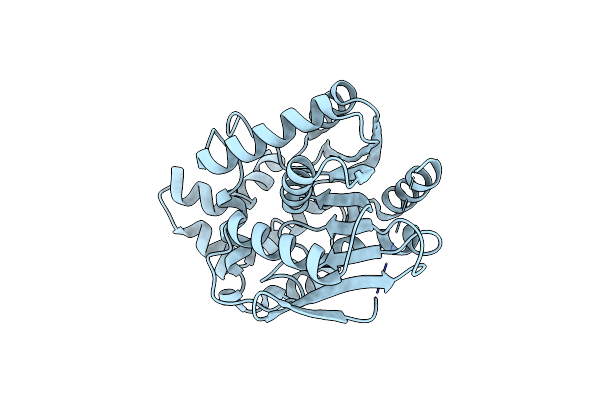
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Circular Permutated At Positions 141-156 (Cphalotagdelta)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: NHE |
 |
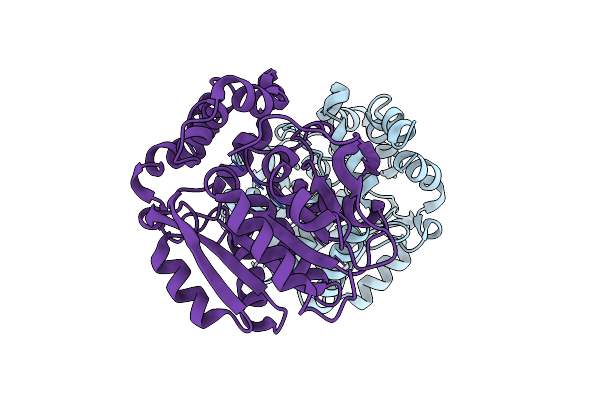
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Circular Permutated At Positions 141-156 (Cphalotagdelta) Fused To M13
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: CL |
 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 Circular Permutated At Positions 154-156 (Cphalotag7_154-156)
Organism: Rhodococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: CL |
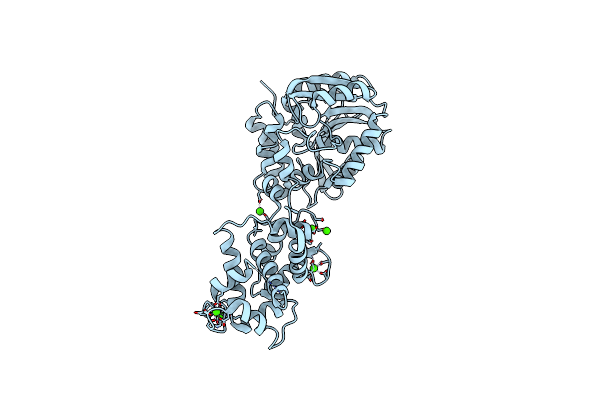 |
X-Ray Structure Of The Haloalkane Dehalogenase Halotag7 With An Insertion Of Calmodulin-M13 Fusion At Position 154-156 That Mimic The Structure Of Caprola, An Calcium Gated Protein Labeling Technology
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-10-11 Classification: HYDROLASE Ligands: CL, CA |
 |