Search Count: 52
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN |
 |
Organism: Thunnus albacares
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: TRANSPORT PROTEIN Ligands: 6WT |
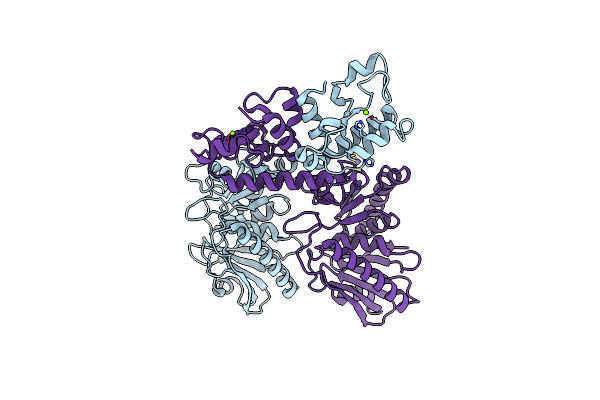 |
Organism: Asticcacaulis sp. ybe204
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2021-11-10 Classification: UNKNOWN FUNCTION Ligands: ZN, MG |
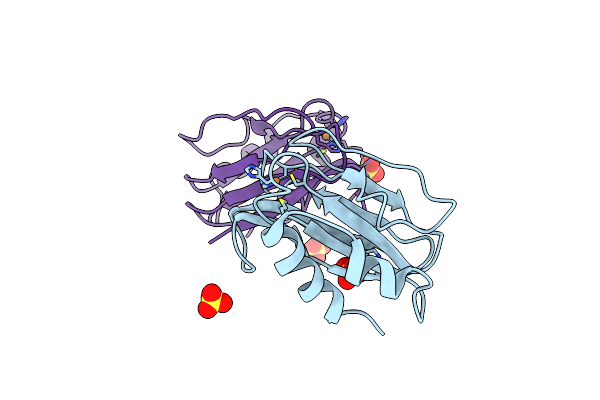 |
Three-Dimensional Structure Of The Mutant K109A Of Paracoccus Pantotrophus Pseudoazurin At Ph 7.0
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2014-07-30 Classification: ELECTRON TRANSPORT Ligands: CU, SO4 |
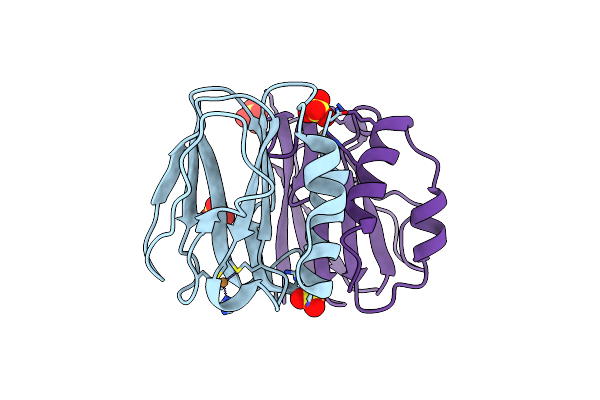 |
Three-Dimensional Structure Of Paracoccus Pantotrophus Pseudoazurin At Ph 6.5
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2014-07-16 Classification: ELECTRON TRANSPORT Ligands: CU, SO4 |
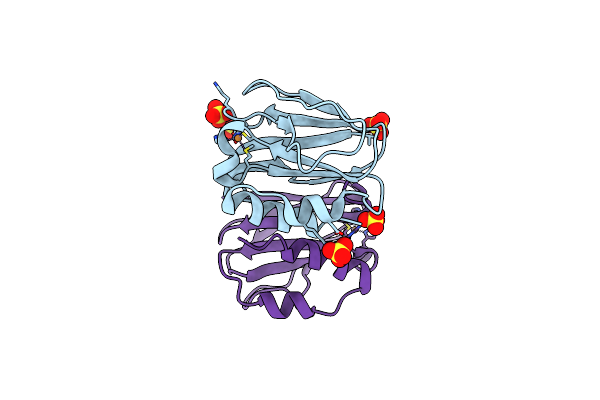 |
Three-Dimensional Structure Of The K109A Mutant Of Paracoccus Pantotrophus Pseudoazurin At Ph 5.5
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2014-07-16 Classification: ELECTRON TRANSPORT Ligands: CU, SO4 |
 |
The Structure Of Cbix, The Terminal Enzyme For Biosynthesis Of Siroheme In Denitrifying Bacteria
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-04-09 Classification: UNKNOWN FUNCTION Ligands: MLT, NA, EDO |
 |
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2009-10-13 Classification: ELECTRON TRANSPORT Ligands: CU, SO4 |
 |
Organism: Paracoccus denitrificans, Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2005-10-06 Classification: OXIDOREDUCTASE Ligands: HEC, ZN |
 |
Organism: Paracoccus pantotrophus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2002-11-28 Classification: OXIDOREDUCTASE Ligands: HEC, DHE, SO4, GOL |
 |
Crystal Structure To 1.74 Angstroms Resolution Of Metmyoglobin From Yellowfin Tuna (Thunnus Albacares): An Example Of A Myoglobin Lacking The D Helix
Organism: Thunnus albacares
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 1993-10-31 Classification: OXYGEN TRANSPORT Ligands: HEM |
 |
 |
 |
 |
 |
 |

