Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
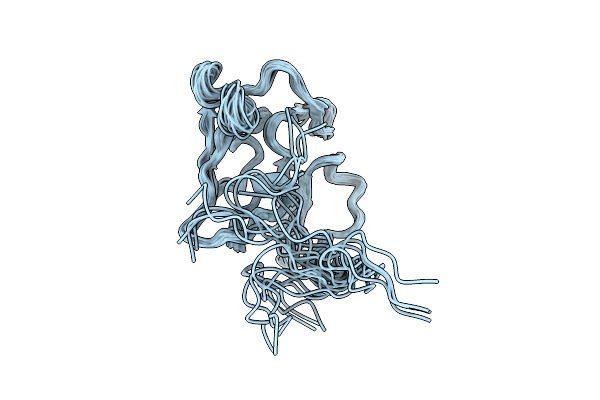 |
Three-Dimensional Structure Of The Merozoite Surface Protein 1 C-Terminal Domain
Organism: Plasmodium berghei
Method: SOLUTION NMR Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN |
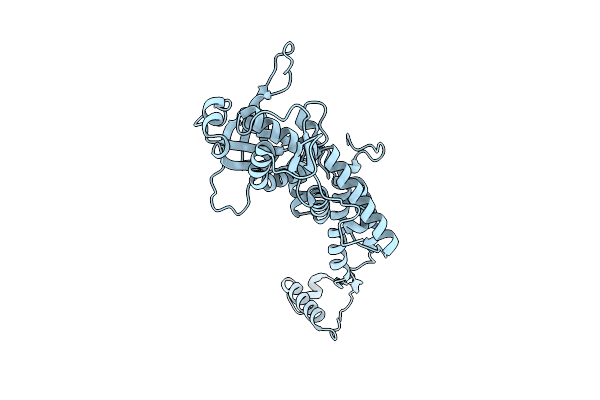 |
Organism: Shigella phage sf11 smd-2017
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRUS |
 |
Crystal Structure Of Ha3 From Clostridium Botulinum Type B With Alpha2,3-Sialyllactose
Organism: Clostridium botulinum b1 str. okra
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN |
 |
Crystal Structure Of Ha3 From Clostridium Botulinum Type B With Alpha2,6-Sialyllactose
Organism: Clostridium botulinum b1 str. okra
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN Ligands: SIA |
 |
Organism: Clostridium phage phicd111
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: ANTIBIOTIC Ligands: ZN |
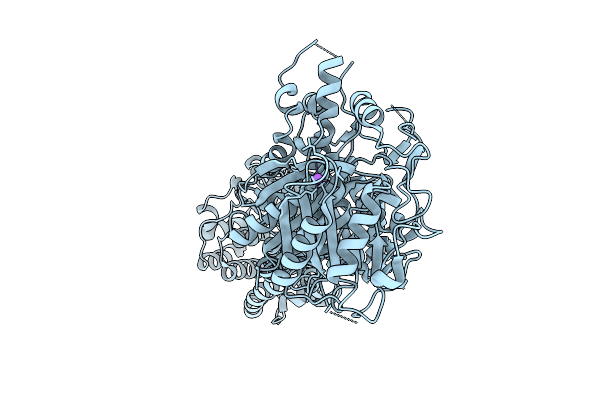 |
Organism: Streptomyces graminofaciens, Synthetic construt
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: TRANSFERASE |
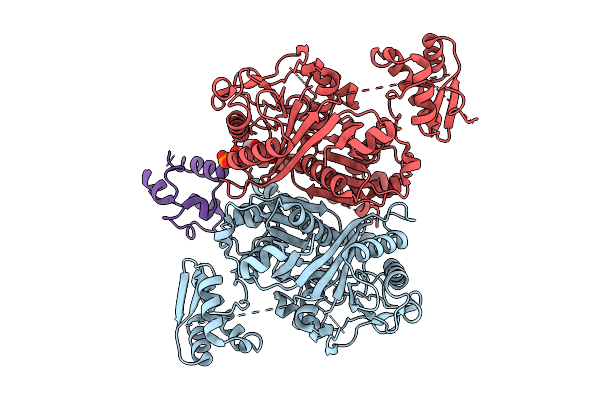 |
Class 3 State Of The Gfsa Ksq-Ancestralat Chimeric Didomain In Complex With The Gfsa Acp Domain
Organism: Streptomyces graminofaciens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LYASE Ligands: 9EF |
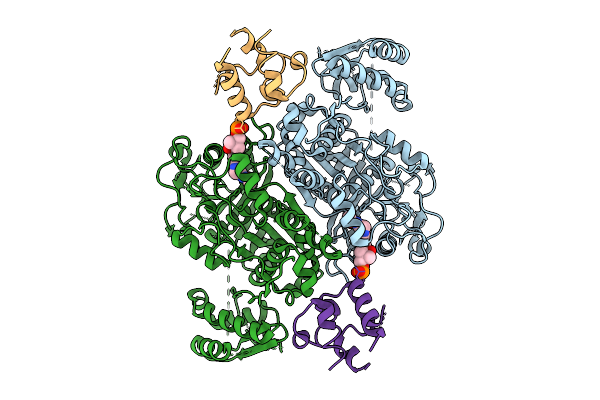 |
Class 1 State Of The Gfsa Ksq-Ancestralat Chimeric Didomain In Complex With The Gfsa Acp Domain
Organism: Streptomyces graminofaciens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LYASE Ligands: 9EF |
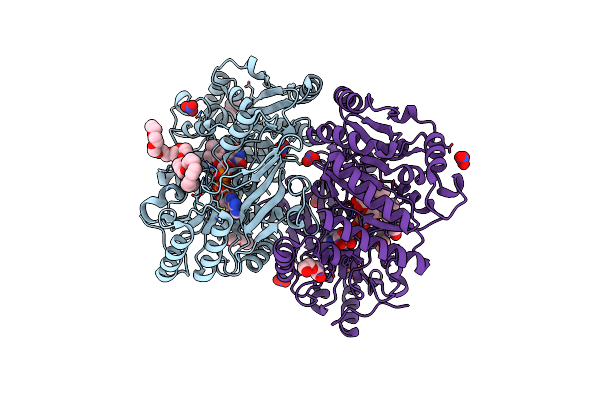 |
Vanillyl-Alcohol Dehydrogenase From Marinicaulis Flavus: P151L Mutant Bound To Eugenol
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, PEG, 1PE, PE4, EOL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, GOL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, PEG |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, CL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, PEG, SO4 |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, CL, PEG |
 |
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-02 Classification: HYDROLASE Ligands: CA, CL, SO4, XHR |
 |
Crystal Structure Of Cytochrome P450 Cyp161H12 From Amycolatopsis Pretoriensis
Organism: Amycolatopsis pretoriensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Norovirus gii.4
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2024-06-19 Classification: VIRAL PROTEIN Ligands: EDO, FMT |
 |
Organism: Norovirus gii.4
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-06-19 Classification: VIRAL PROTEIN Ligands: EDO, FMT |