Search Count: 544
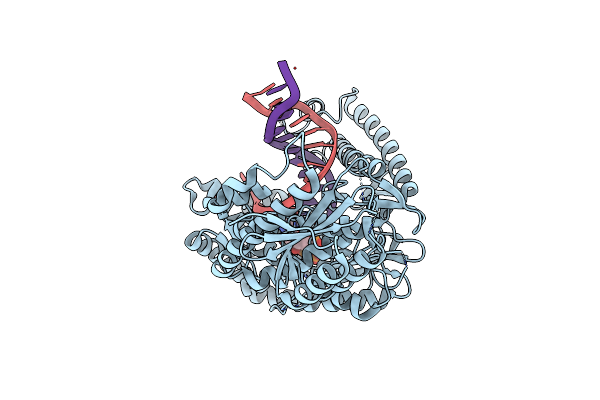 |
Structure Of Lates Calcarifer Dna Polymerase Theta Polymerase Domain With Long Duplex Dna, Complex Ia
Organism: Lates calcarifer
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: DNA BINDING PROTEIN/DNA Ligands: DGT, MG |
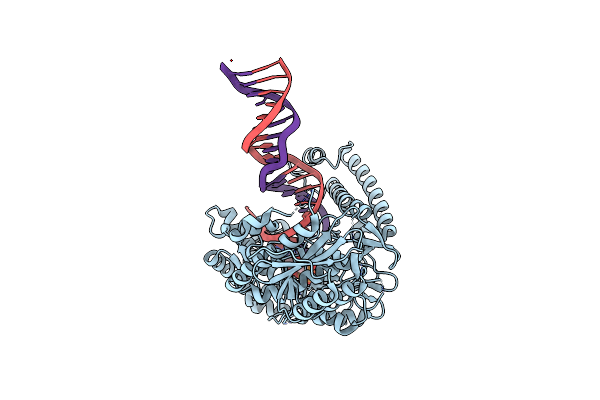 |
Structure Of Lates Calcarifer Dna Polymerase Theta Polymerase Domain With Long Duplex Dna, Complex Ia
Organism: Lates calcarifer
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: DNA BINDING PROTEIN Ligands: DGT, MG |
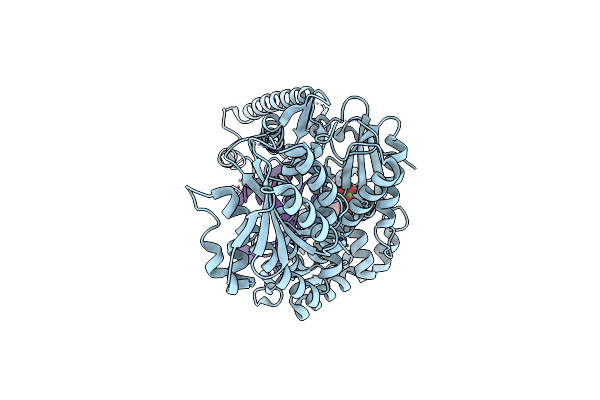 |
Structure Of Lates Calcarifer Dna Polymerase Theta Polymerase Domain With Hairpin Dna
Organism: Lates calcarifer
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: DNA BINDING PROTEIN/DNA Ligands: MG, DDS |
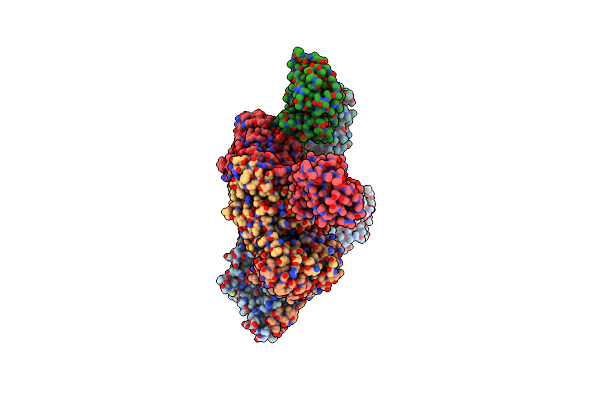 |
Organism: Lates calcarifer, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: REPLICATION/DNA Ligands: ATP, MG |
 |
Crystal Structure Of Arginine Kinase From The Spider Polybetes Pythagoricus In Complex With Arginine
Organism: Polybetes pythagoricus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-09-06 Classification: TRANSFERASE Ligands: ARG, NA |
 |
Crystal Structure Of Substrate-Free Arginine Kinase From Spider Polybetes Pythagoricus
Organism: Polybetes pythagoricus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2017-08-30 Classification: TRANSFERASE |
 |
Organism: Streptomyces cirratus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2014-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, GOL |
 |
Organism: Streptomyces cirratus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2014-11-19 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, AMP, GOL |
 |
Crystal Structure Of Lepb Gap Domain From Legionella Drancourtii In Complex With Rab1-Gdp And Alf3
Organism: Legionella drancourtii, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2013-05-08 Classification: HYDROLASE ACTIVATOR/PROTEIN TRANSPORT Ligands: ACY, GDP, AF3, MG |
 |
Biochemical And Structural Characterization Of The Mpaa Amidase As Part Of A Conserved Scavenging Pathway For Peptidoglycan Derived Peptides In Gamma-Proteobacteria
Organism: Vibrio campbellii atcc baa-1116
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2012-09-26 Classification: HYDROLASE Ligands: ZN, EDO |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

