Search Count: 2,160
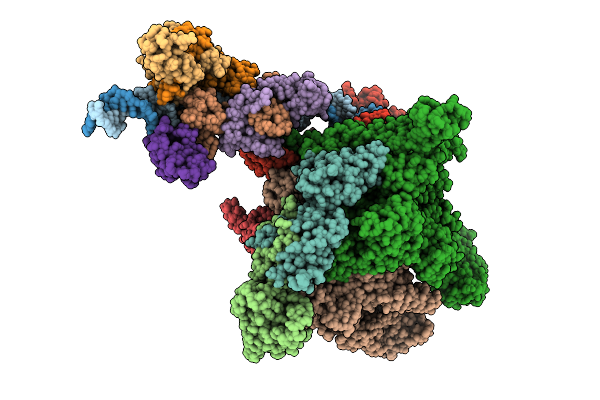 |
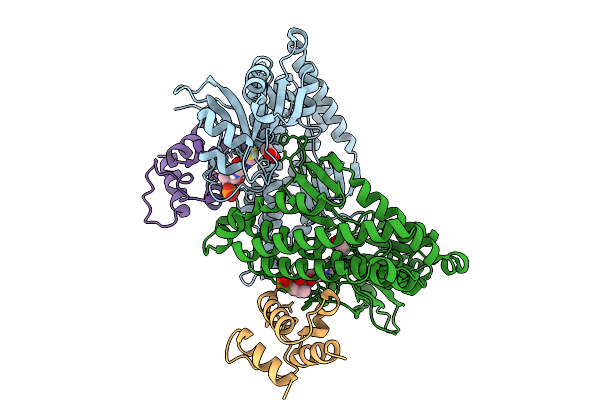
Cryo-Em Structure Of E.Coli Transcription Initiation Complex With Escherichia Phage Mu Late Transcription Activator C
Organism: Escherichia coli (strain k12), Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION/DNA |
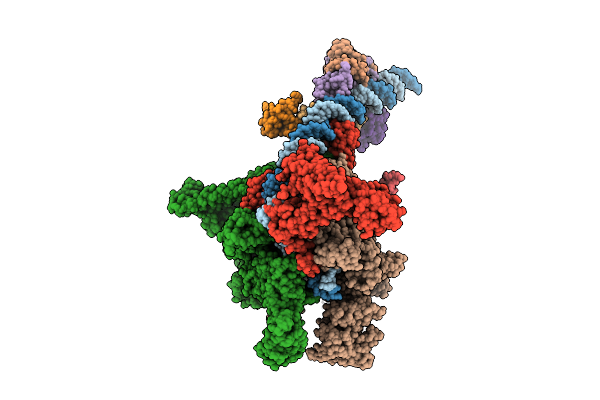 |
Cryo-Em Structure Of E.Coli Transcription Initiation Complex With Escherichia Phage Mu Middle Transcription Activator Mor
Organism: Escherichia coli (strain k12), Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION/DNA |
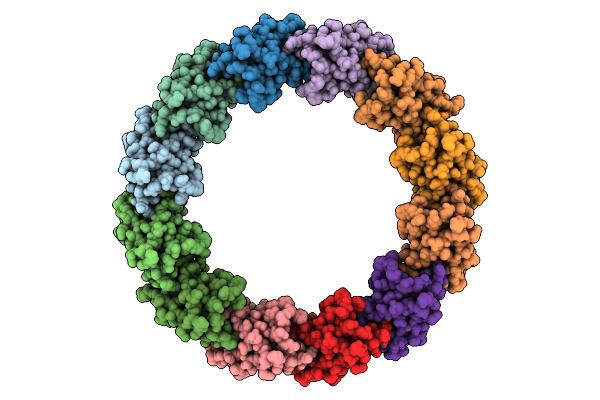 |
Organism: Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN |
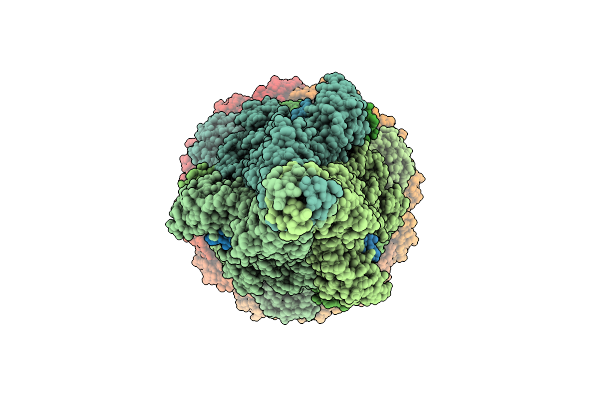 |
Organism: Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: SF4 |
 |
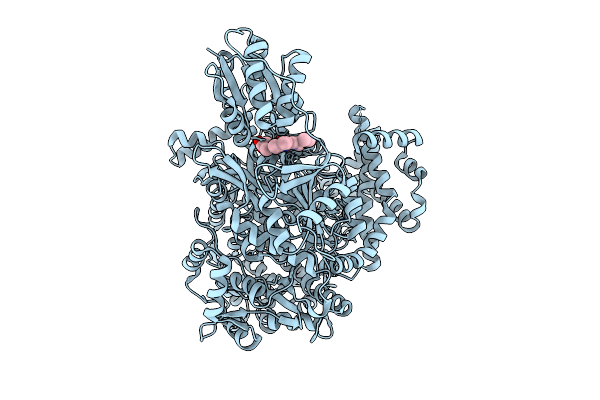
Organism: Streptomyces caelestis
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: BIOSYNTHETIC PROTEIN Ligands: ACT, PNS |
 |
Crystal Structure Of Ccbd C19S Mutant Complexed With Single Pcp Domain Of Ccbz
Organism: Streptomyces caelestis
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: BIOSYNTHETIC PROTEIN Ligands: MLI, PNS |
 |
Organism: Bacteroides
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: METAL TRANSPORT Ligands: HEM, NA |
 |
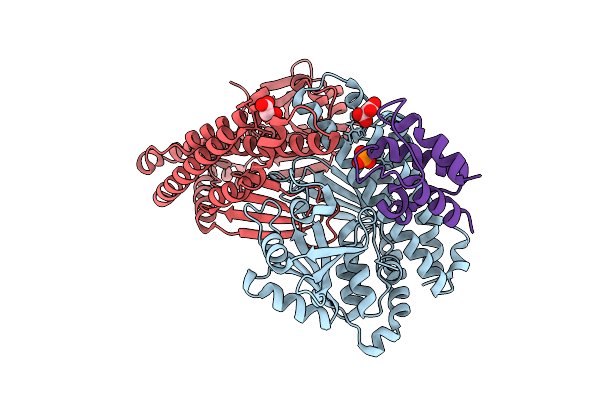
Distinct Quaternary States, Intermediates, And Autoinhibition During Loading Of The Dnab-Replicative Helicase By The Phage Lambda P Helicase Loader
Organism: Escherichia phage lambda
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: REPLICATION |
 |
Ecoli Dnab Helicase And Phage Lambda Loader P With Adp-Mg In A 6:5 Stoichiometry Ratio.
Organism: Escherichia coli, Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN Ligands: ADP, MG |
 |
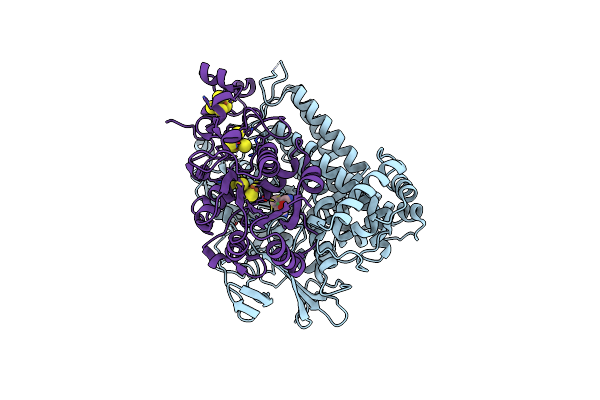
Ecoli Dnab Helicase And Phage Lambda Loader P With Adp-Mg In A 6:6 Stoichiometry Ratio.
Organism: Escherichia coli, Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN Ligands: ADP, MG |
 |
Crystal Structure Of The C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The Air-Oxidized State At 1.93 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, 35L, SF3, CL |
 |
Organism: Metagenomes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: MLA, BGC, NHE |
 |
Organism: Metagenomes
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: NHE, NA, MLA |
 |
Crystal Structure Of Gh1 Beta-Glucosidase Td2F2 E352Q Laminaribiose Complex
Organism: Metagenomes
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: MLA, NHE, SO4 |
 |
Crystal Structure Of The C19G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.6 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, NA, PO4, SF4, F3S, CL, ER2 |
 |
Crystal Structure Of The C120G Variant Of The Membrane-Bound [Nife]-Hydrogenase From Cupriavidus Necator In The H2-Reduced State At 1.65 A Resolution.
Organism: Cupriavidus necator h16
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-09-18 Classification: OXIDOREDUCTASE Ligands: NFU, MG, CL, SF4, F3S, 35L, F4S |
 |
E.Coli Gyrase Holocomplex With Cleaved Chirally Wrapped 217 Bp Dna Fragment And Moxifloxacin
Organism: Escherichia coli, Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: ISOMERASE Ligands: MG, MFX |
 |
Organism: Escherichia coli, Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: ISOMERASE Ligands: MG |
 |
Organism: Streptomyces caelestis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-08-14 Classification: LYASE |
 |
Organism: Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRAL PROTEIN/DNA Ligands: ATP |

