Search Count: 71
 |
Organism: Acidothermus cellulolyticus
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: DNA BINDING PROTEIN/DNA/RNA |
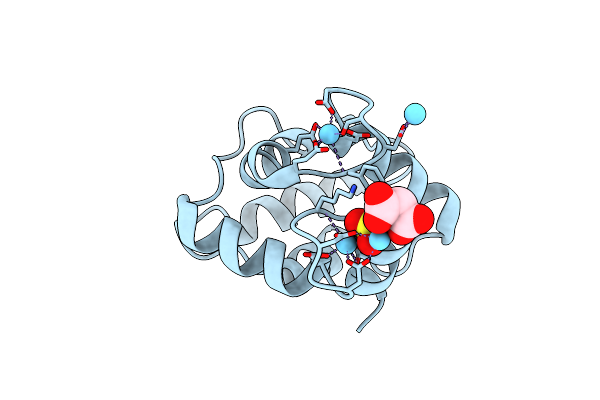 |
Organism: Gadus morhua
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-12-04 Classification: ALLERGEN Ligands: LA, MLI, CL, SO4 |
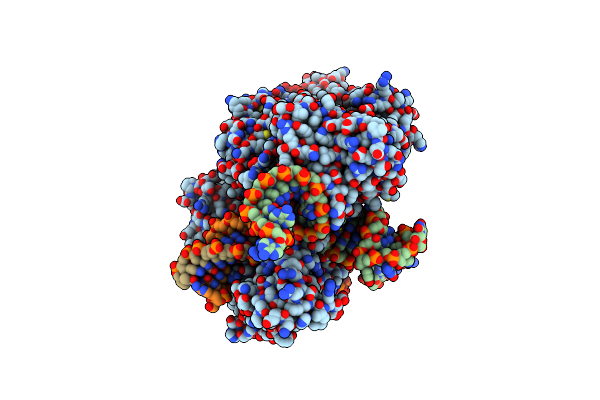 |
Structure Of Acidothermus Cellulolyticus Cas9 Ternary Complex (Cleavage Intermediate 2)
Organism: Acidothermus cellulolyticus 11b, Acidothermus cellulolyticus
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Organism: Sin nombre orthohantavirus
Method: ELECTRON MICROSCOPY Release Date: 2023-07-19 Classification: VIRAL PROTEIN |
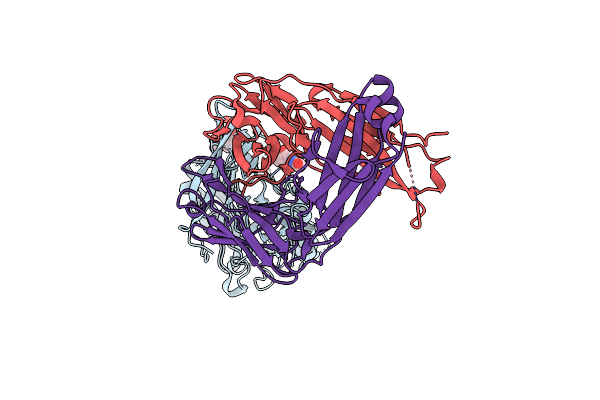 |
Organism: Sin nombre orthohantavirus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-24 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-05-04 Classification: HYDROLASE |
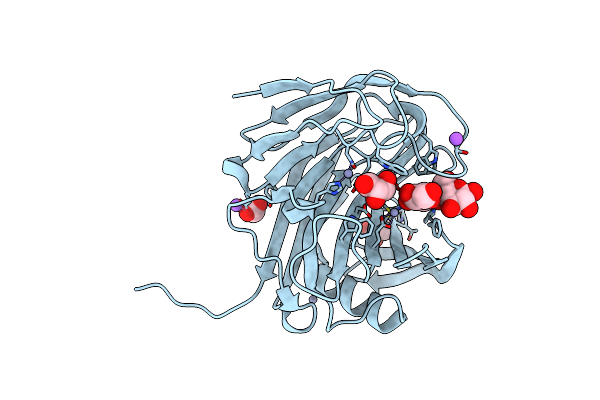 |
Crystal Structure Of The Gh12 Domain From Acidothermus Cellulolyticus Guxa Bound To Cellobiose
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-05-04 Classification: HYDROLASE Ligands: ZN, CL, NA, ACT, GOL |
 |
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2021-12-01 Classification: HYDROLASE Ligands: PGE, SCN, PEG, EDO |
 |
Organism: Acidothermus cellulolyticus, Acidothermus cellulolyticus (strain atcc 43068 / 11b)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-12-01 Classification: HYDROLASE Ligands: MG, EDO, ACP, ACT, PEG, PGE |
 |
Crystal Structure Of Depupylase Dop In Complex With Phosphorylated Pup And Adp
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2021-12-01 Classification: HYDROLASE Ligands: K, MG, EDO, ADP, ACT, PEG, PGE |
 |
Crystal Structure Of Depupylase Dop In Complex With Pup And Adp/Trifluoromagnesate
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Release Date: 2021-12-01 Classification: HYDROLASE Ligands: MG, ADP, EDO, PEG, ACT, PGE, KQB |
 |
Crystal Structure Of Depupylase Dop In Complex With Pup And Adp/Tetrafluoromagnesate
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-12-01 Classification: HYDROLASE Ligands: MG, ADP, EDO, PEG, ACT, PGE, SO4, MF4 |
 |
Atomic Model Of Andes Virus Glycoprotein Spike Tetramer Generated By Fitting Into A Tula Virus Reconstruction
Organism: Tula orthohantavirus
Method: ELECTRON MICROSCOPY Release Date: 2020-10-14 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structure, Dynamics And Stability Of Allergen Cod Parvalbumin Gad M 1 By Solution And High-Pressure Nmr.
Organism: Gadus morhua
Method: SOLUTION NMR Release Date: 2014-08-20 Classification: CONTRACTILE PROTEIN Ligands: CA |
 |
Crystal Structure Of Uracil-Dna Glycosylase From Cod (Gadus Morhua) In Complex With The Proteinaceous Inhibitor Ugi
Organism: Gadus morhua, Bacillus phage pbs2
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2014-08-13 Classification: HYDROLASE/HYDROLASE INHIBITOR |
 |
The Crystal Structure Of G-Type Lysozyme From Atlantic Cod (Gadus Morhua L.) In Complex With Nag Oligomers Sheds New Light On Substrate Binding And The Catalytic Mechanism. Native Structure To 1.9
Organism: Gadus morhua
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-10-20 Classification: HYDROLASE Ligands: CO |
 |
The Crystal Structure Of G-Type Lysozyme From Atlantic Cod (Gadus Morhua L.) In Complex With Nag Oligomers Sheds New Light On Substrate Binding And The Catalytic Mechanism. Structure With Nag To 1.7
Organism: Gadus morhua
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-10-20 Classification: HYDROLASE |
 |
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-07-05 Classification: HYDROLASE |
 |
Crystal Structure Of Uracil-Dna Glycosylase From Atlantic Cod (Gadus Morhua)
Organism: Gadus morhua
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-04-05 Classification: HYDROLASE Ligands: GOL, CL |
 |
Acidothermus Cellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1996-10-14 Classification: GLYCOSYL HYDROLASE |

