Search Count: 164
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: SO4, ACT, GOL, CL, NA |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: GOL |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CL, GOL, ACT |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CL, A1EGH, ACT, PO4 |
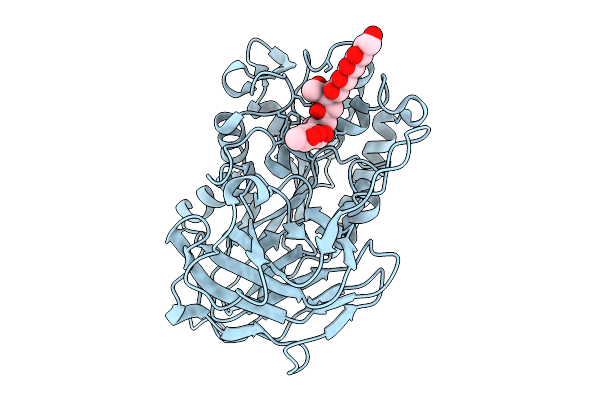 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: A1EGL, CL |
 |
Organism: Sabia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: VIRAL PROTEIN Ligands: NAG, ZN |
 |
Organism: Sabia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: VIRAL PROTEIN Ligands: NAG, K |
 |
Structure Of The Sabia Virus Spike Complex H157M Mutant In A Closed Conformation
Organism: Sabia virus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: VIRAL PROTEIN Ligands: NAG, ZN |
 |
Organism: Halalkalicoccus jeotgali b3
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2021-04-14 Classification: TRANSFERASE |
 |
Organism: Halalkalicoccus jeotgali b3
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-04-14 Classification: TRANSFERASE |
 |
Organism: Halalkalicoccus jeotgali b3
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2021-04-14 Classification: TRANSFERASE |
 |
Solution Structure Of Zalpha Domain Of Goldfish Zbp-Containing Protein Kinase
Organism: Carassius auratus
Method: SOLUTION NMR Release Date: 2016-02-03 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Zalpha Domain From Carassius Auratus Pkz In Complex With Z-Dna
Organism: Carassius auratus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-07-03 Classification: TRANSFERASE/DNA Ligands: MN |
 |
 |
 |
 |
Organism: Hieroxestinae sp. BIOUG20573-B10
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
NA
|
 |
NA
|
 |
NA
|

