Search Count: 22,950
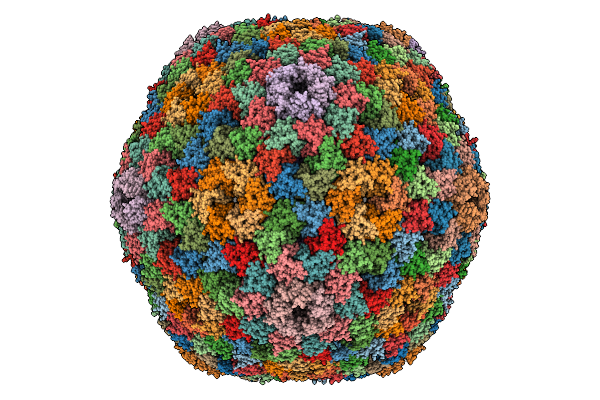 |
Organism: Halothiobacillus neapolitanus c2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: STRUCTURAL PROTEIN |
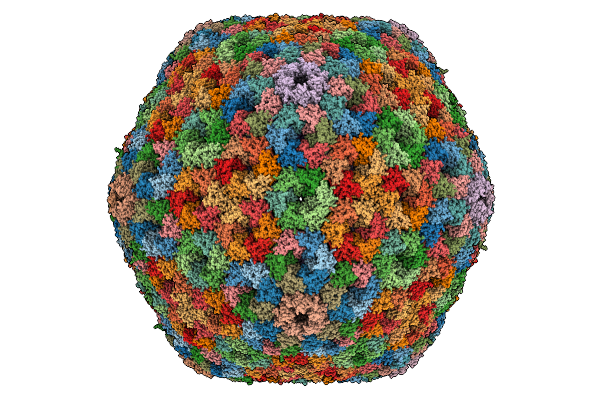 |
Cryo-Em Structure Of Carboxysomal Mid-Shell: T = 16 Shell Under C1 Symmetry.
Organism: Halothiobacillus neapolitanus c2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: STRUCTURAL PROTEIN |
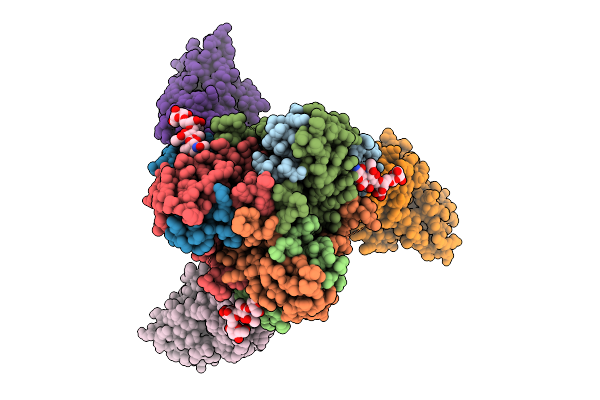 |
Organism: Homo sapiens, Ravn virus - ravn, kenya, 1987
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Organism: Ravn virus - ravn, kenya, 1987
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Organism: Vicugna pacos, Ravn virus - ravn, kenya, 1987
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
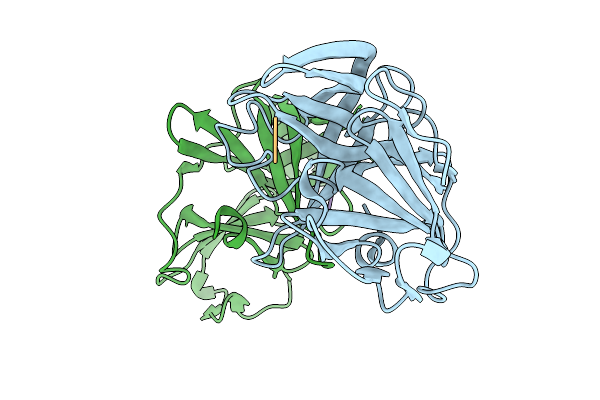 |
Organism: Norovirus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Thermothielavioides terrestris (strain atcc 38088 / nrrl 8126)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, MG |
 |
Crystal Structure Of Xanthobacter Autotrophicus Sparda Mutant Lacking Dren Nuclease Domains
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: ACY, GOL, TRS, PEG, CL, BME, TAR |
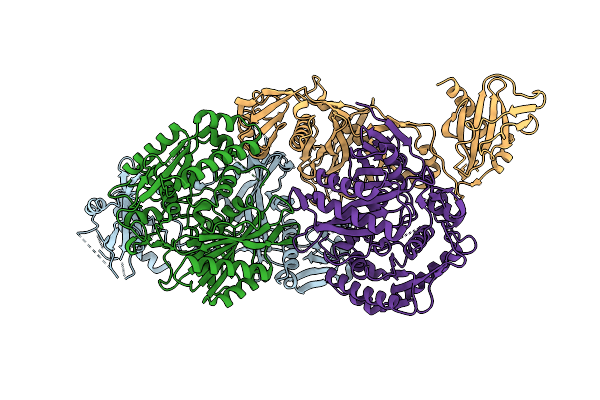 |
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: SO4 |
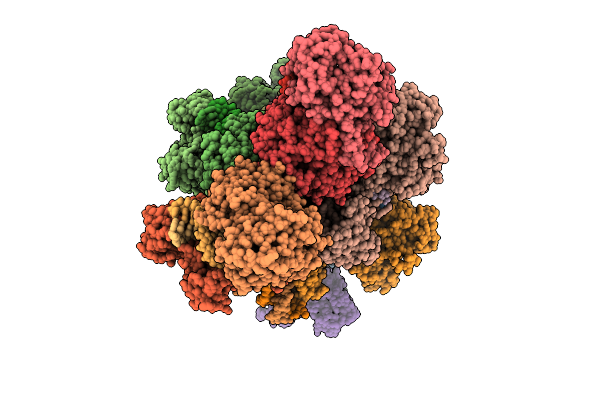 |
Organism: Xanthobacter autotrophicus py2, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: CA |
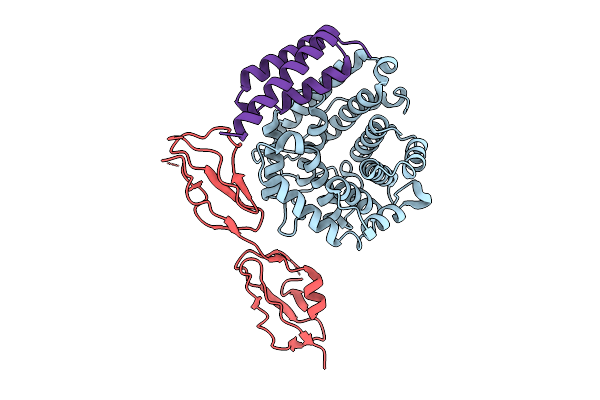 |
Organism: Homo sapiens, Staphylococcus aureus subsp. aureus mu50
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: IMMUNE SYSTEM |
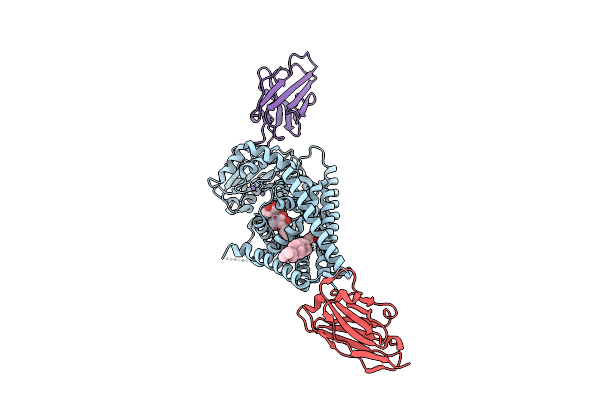 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: Y01, MN, LMT |
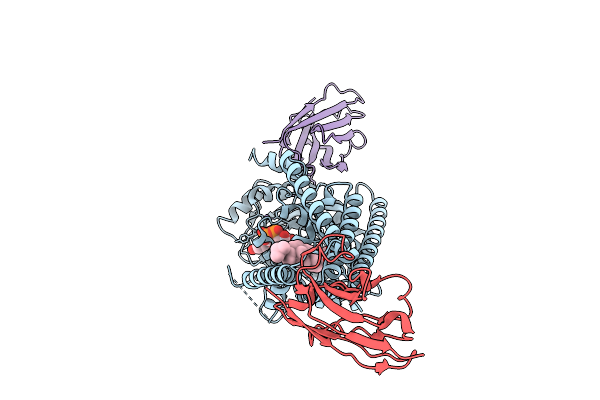 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: UGA, MN, Y01 |
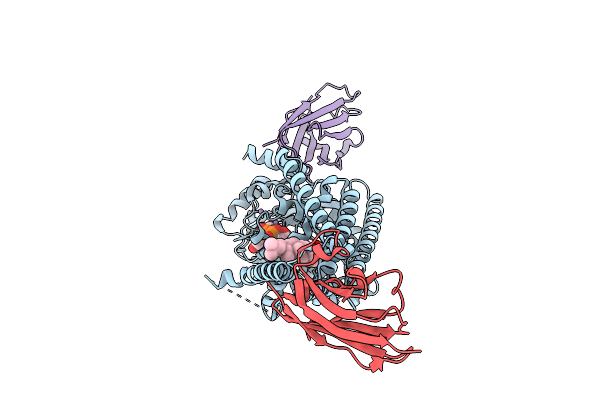 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: UGA, Y01, MN |
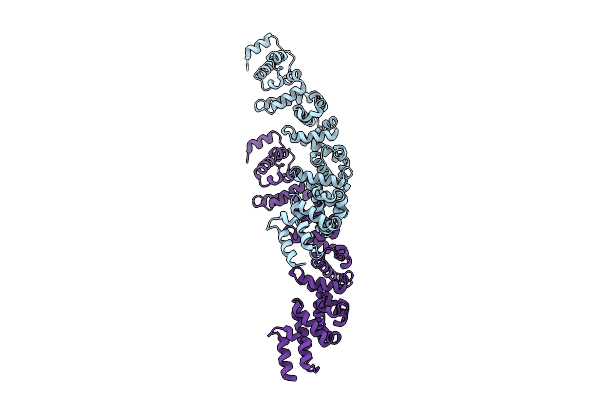 |
Organism: Phytophthora capsici
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: CYTOSOLIC PROTEIN |
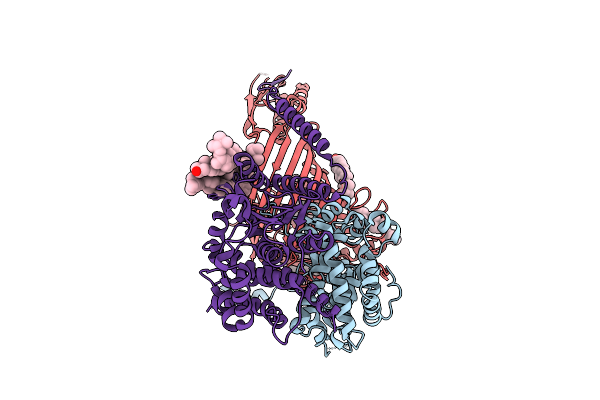 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
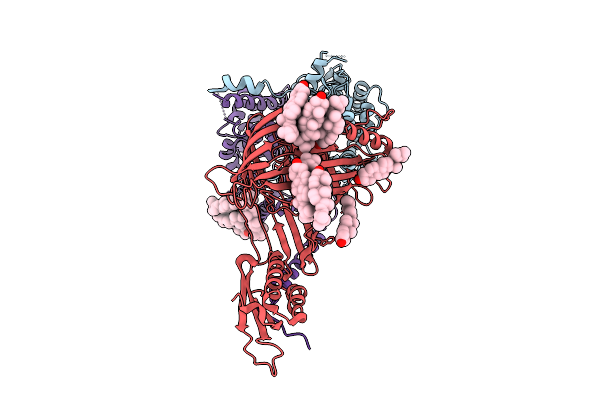 |
Organism: Thermothelomyces thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: ERG |
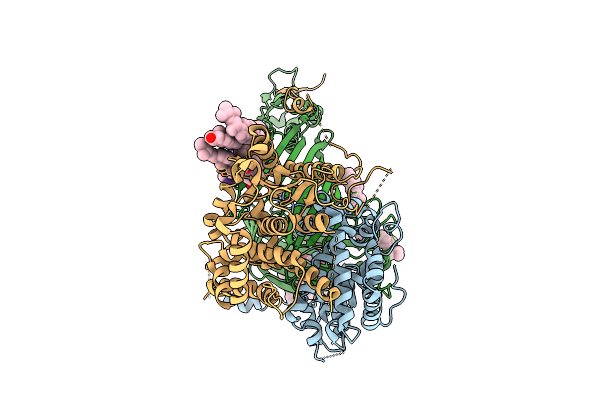 |
Organism: Thermothelomyces thermophilus, Photorhabdus khanii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN/ANTIBIOTIC Ligands: ERG |
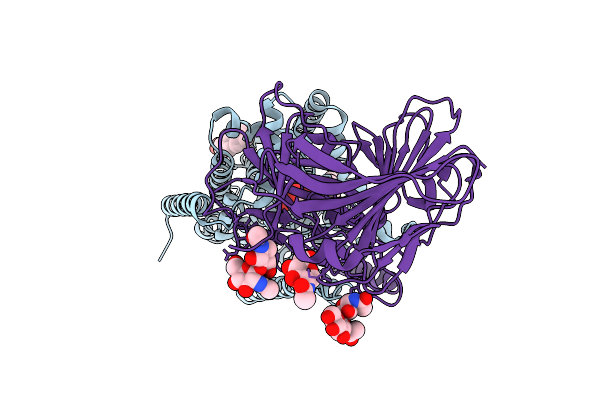 |
Cryo-Em Structure Of Candida Glabrata Gpi Mannosyltransferase I Bound To Dol-P-Man
Organism: Nakaseomyces glabratus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN Ligands: MJC, Y01, NAG |
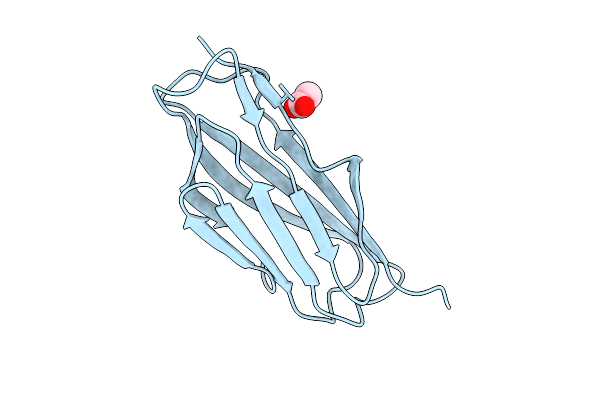 |
Organism: Acinetobacter silvestris
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: CELL ADHESION Ligands: ACT |

