Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
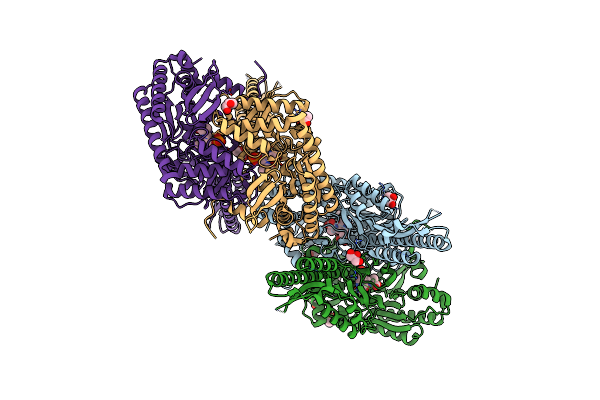
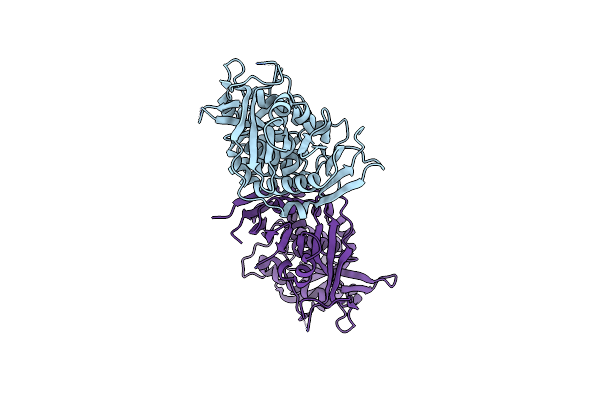
Glycosyltransferase C From The Limosilactobacillus Reuteri Accessory Secretion System. Complex With Udp-Glcnac.
Organism: Limosilactobacillus
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: TRANSFERASE Ligands: UDP, CIT, GOL, EPE, CL, UD1 |
 |
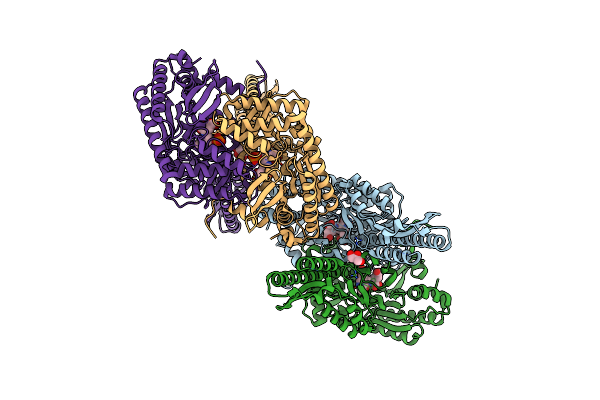
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In Complex With Amino Donor L-Ala
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-02-05 Classification: TRANSFERASE Ligands: F0G, GOL, ACY |
 |
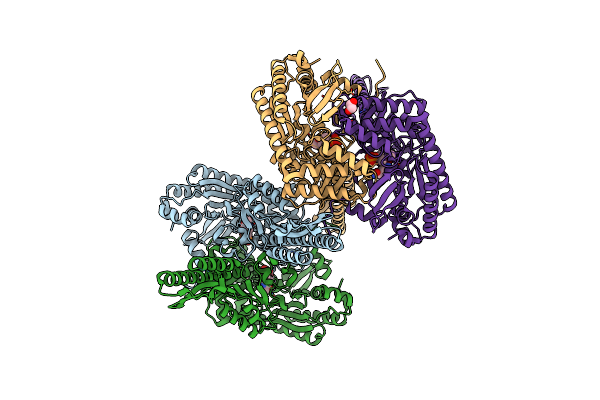
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In Complex With Amino Donor L-Glu
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-02-05 Classification: TRANSFERASE Ligands: PL6, GOL |
 |
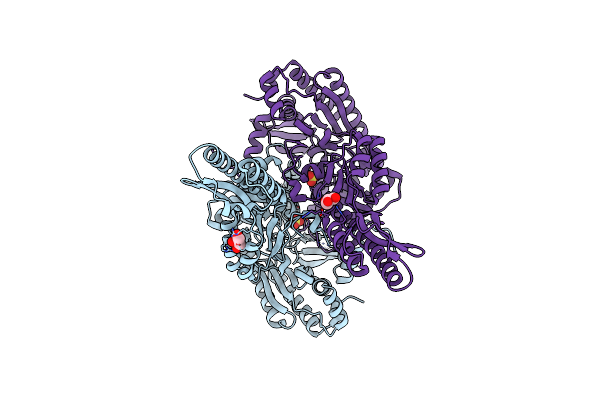
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In Complex With Amino Donor L-Gln
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-02-05 Classification: TRANSFERASE Ligands: PLP, ACY, GOL, GLN |
 |
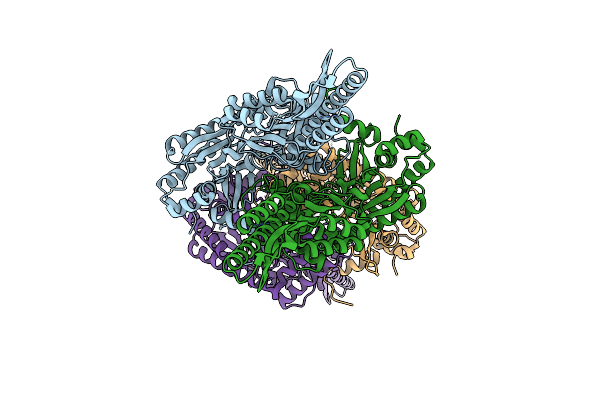
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In I222 Space Group
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-02-05 Classification: TRANSFERASE Ligands: GOL, CL, SO4 |
 |
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In C2 Space Group
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-02-05 Classification: TRANSFERASE |
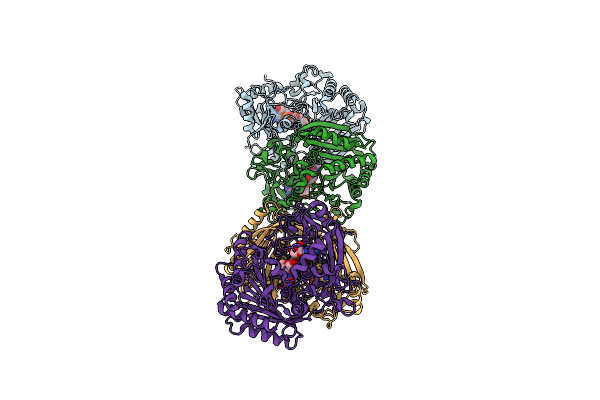 |
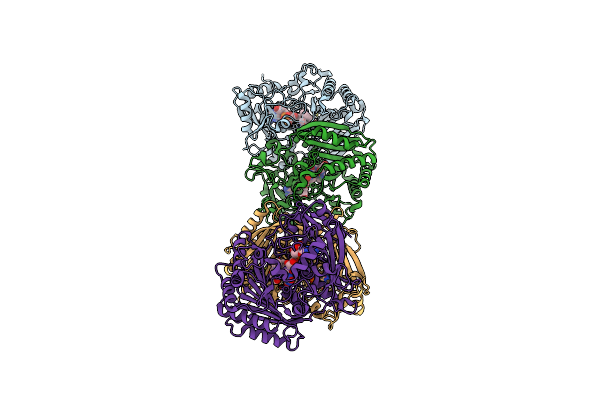
Crystal Structure Of Crmk, A Flavoenzyme Involved In The Shunt Product Recycling Mechanism In Caerulomycin Biosynthesis
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
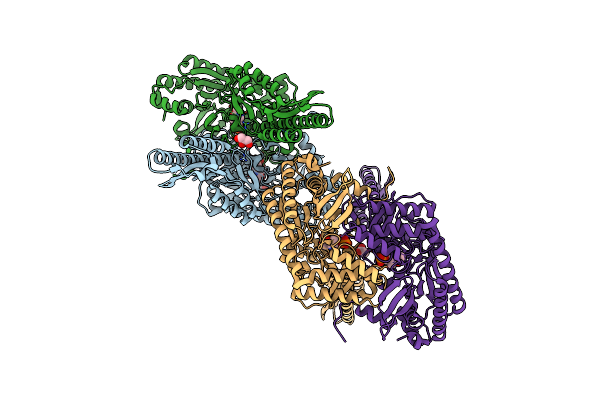
Crystal Structure Of Crmk, A Flavoenzyme Involved In The Shunt Product Recycling Mechanism In Caerulomycin Biosynthesis
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: FAD, 67L, 67K |
 |
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In Complex With Plp
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: PLP, GOL, ACY |
 |
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In Complex With Pmp
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: PMP, GOL, PGE |
 |
Crystal Structure Of Aminotransferase Crmg From Actinoalloteichus Sp. Wh1-2216-6 In Complex With The Pmp External Aldimine Adduct With Caerulomycin M
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-08-10 Classification: TRANSFERASE Ligands: 5B6, GOL |
 |
Structure Of Eukaryotic Translation Initiation Factor 3 Subunit D (Eif3D) Cap Binding Domain From Nasonia Vitripennis, Crystal Form 1
Organism: Nasonia vitripennis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-07-27 Classification: TRANSLATION Ligands: CL |
 |
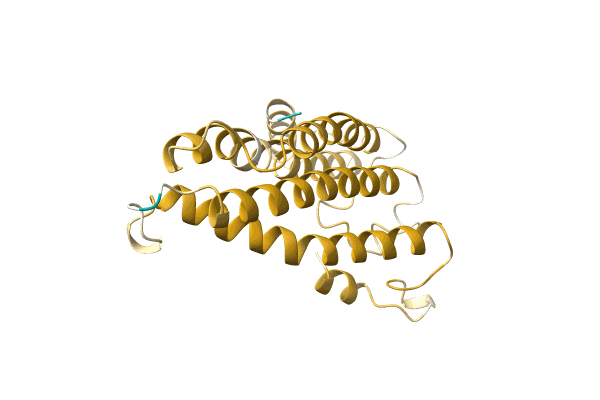
Structure Of Eukaryotic Translation Initiation Factor 3 Subunit D (Eif3D) Cap Binding Domain From Nasonia Vitripennis, Crystal Form 2
Organism: Nasonia vitripennis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-07-27 Classification: TRANSLATION Ligands: GOL |
 |
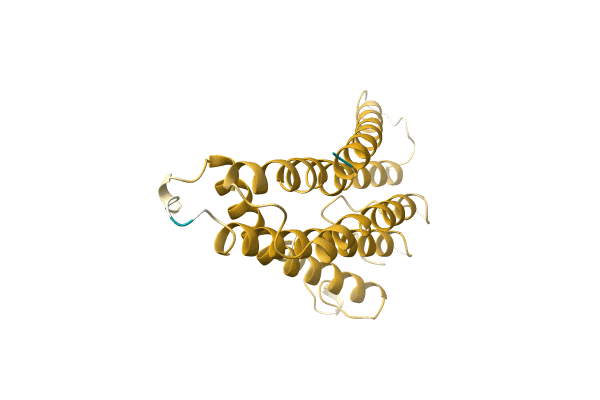
Structure Of Eukaryotic Translation Initiation Factor 3 Subunit D (Eif3D) Cap Binding Domain From Nasonia Vitripennis, Crystal Form 3
Organism: Nasonia vitripennis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-07-27 Classification: TRANSLATION |
 |
 |
 |
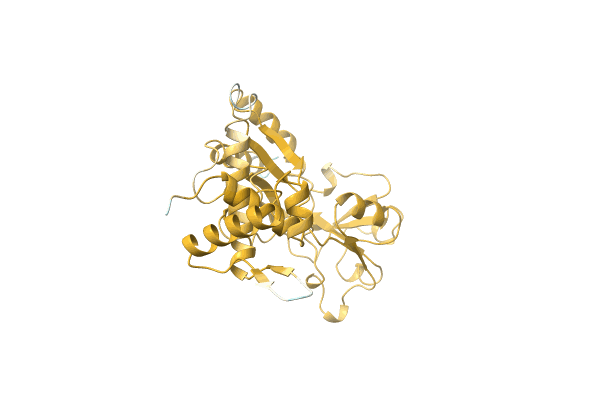
Organism: Dichomeridinae sp. BIOUG20274-F03
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
 |
 |