Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
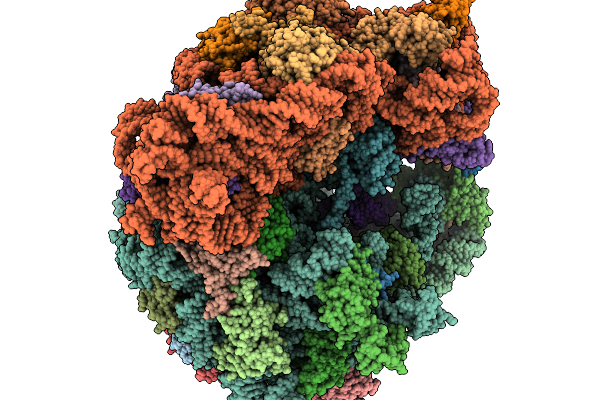 |
Structure Of Clim-Stalled Bacillus Subtilis 70S Ribosome With Release Factor Bound In The A-Site
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME Ligands: ZN |
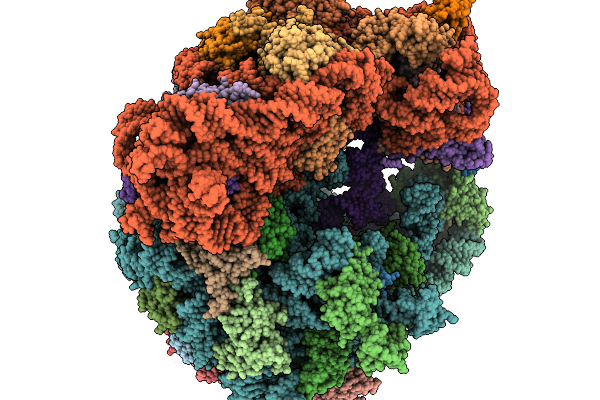 |
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME |
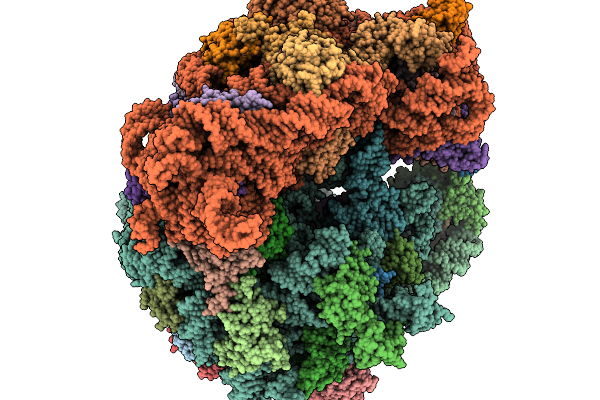 |
Structure Of Clim-Stalled Bacillus Subtilis 70S Ribosome With Trna-Tyr In The A-Site
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME Ligands: ZN |
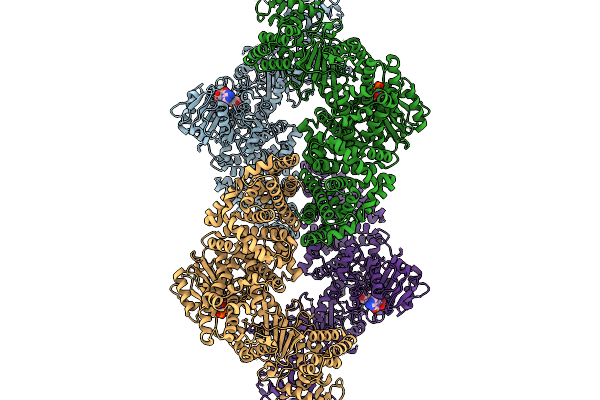 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Open Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
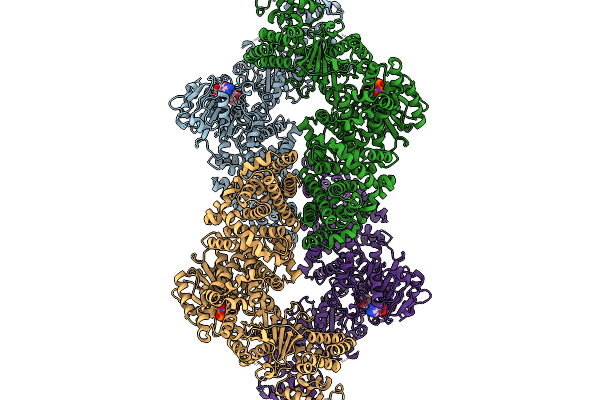 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed1 Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
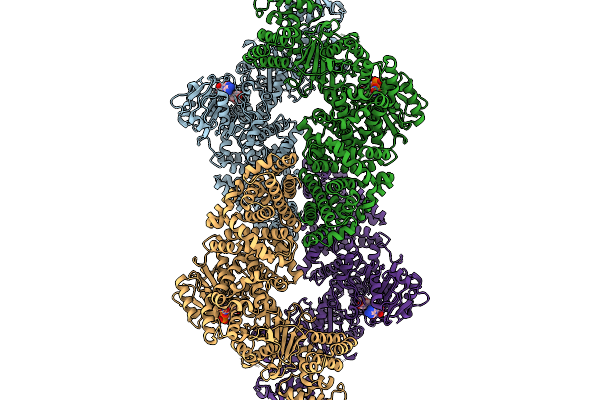 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed2 Tetramer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Empty Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE |
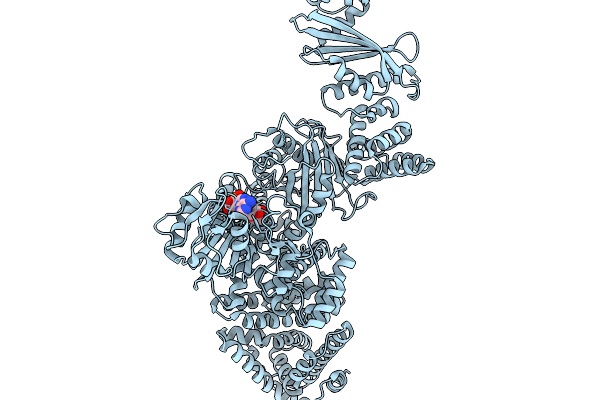 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor-Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
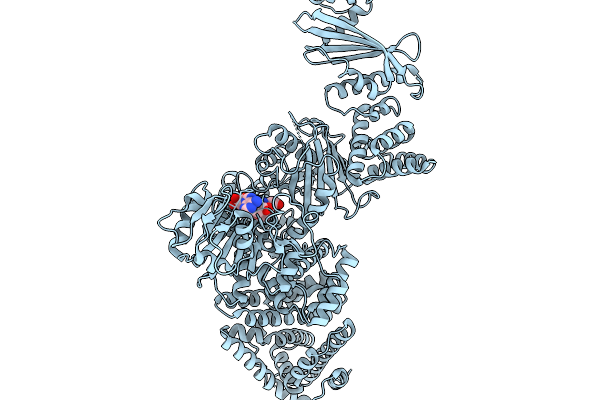 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor/Ligand-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: GLU, NAD |
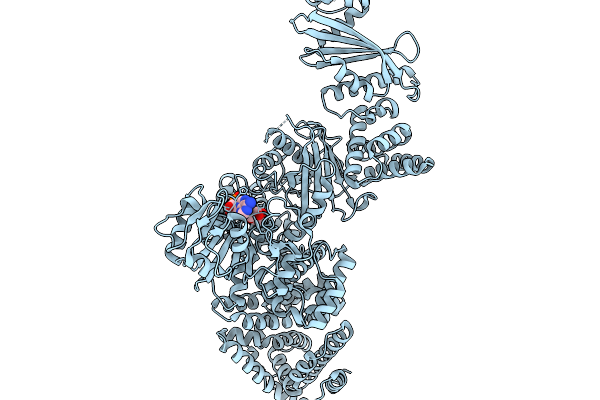 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Total-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of The Coiled-Coil Based Fibril Core Of The Tardigrade Cahs-8 Protein
Organism: Hypsibius exemplaris
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: PROTEIN FIBRIL |
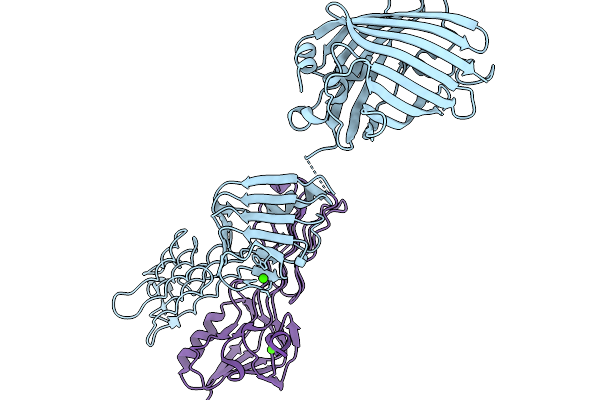 |
Organism: Aequorea victoria, Escherichia phage pp01
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: CELL ADHESION Ligands: CA |
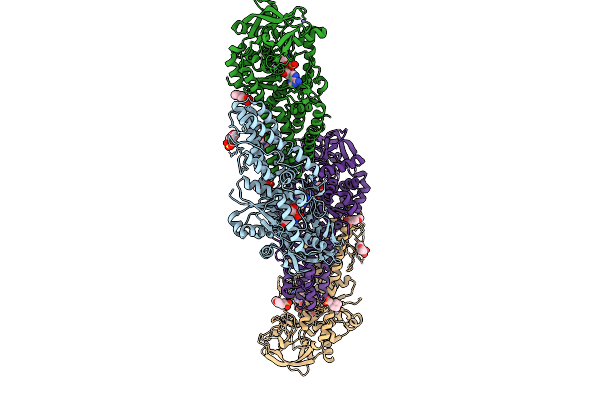 |
Organism: Streptomyces candidus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: LIGASE Ligands: MES, ZN, GSU |
 |
Organism: Respiratory syncytial virus a2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Respiratory syncytial virus a2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Organism: Respiratory syncytial virus a2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
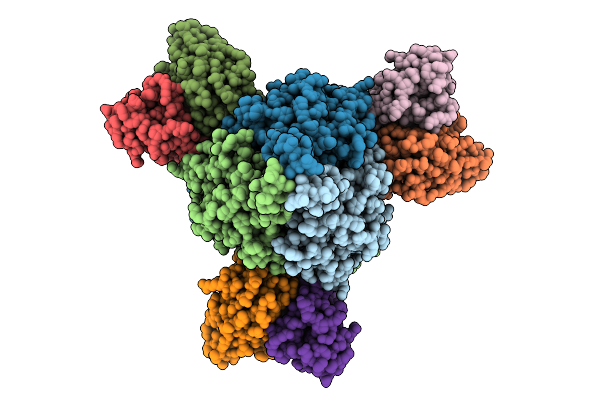 |
Organism: Respiratory syncytial virus a2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Respiratory syncytial virus a2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Respiratory syncytial virus a2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Respiratory syncytial virus a2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN |