Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Pseudomonas phage pap2
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: 4UR |
 |
Organism: Pseudomonas phage pap2
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: 3AM |
 |
Organism: Pseudomonas phage pap2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: 3AM, 3GP |
 |
Organism: Pseudomonas phage pap2
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: EDO, 3AM, 3GP |
 |
Organism: Pseudomonas phage pap2
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: GOL, EDO, 1SY |
 |
Organism: Pseudomonas phage pap2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-02-22 Classification: VIRAL PROTEIN Ligands: 4BW |
 |
Organism: Pseudomonas phage pap2
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2023-02-22 Classification: VIRAL PROTEIN Ligands: EDO, GOL |
 |
Organism: Pseudomonas phage pap2
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-02-22 Classification: VIRAL PROTEIN Ligands: 2BA, EDO |
 |
Organism: Tenebrio molitor
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2012-02-22 Classification: HYDROLASE Ligands: PO4, PG4, PG6 |
 |
Structure Of Digestive Procathepsin L2 Proteinase From Tenebrio Molitor Larval Midgut
Organism: Tenebrio molitor
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-02-01 Classification: HYDROLASE Ligands: ACT |
 |
Crystal Structure Of Serpin48, Which Is A Highly Specific Serpin In The Insect Tenebrio Molitor
Organism: Tenebrio molitor
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-10-27 Classification: HYDROLASE INHIBITOR |
 |
Organism: Tenebrio molitor
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-09-26 Classification: ISOMERASE Ligands: TRS |
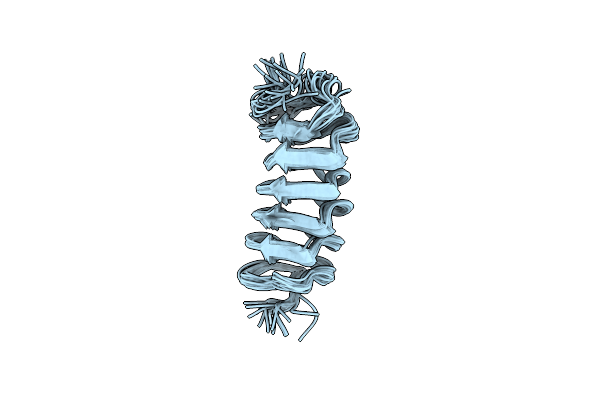 |
Organism: Tenebrio molitor
Method: SOLUTION NMR Release Date: 2002-05-22 Classification: ANTIFREEZE PROTEIN |
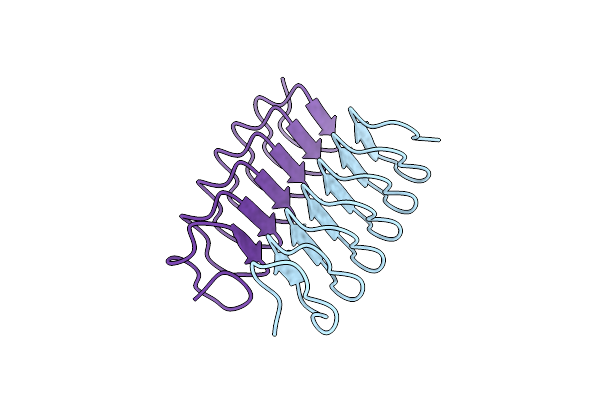 |
Organism: Tenebrio molitor
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2000-08-09 Classification: ANTIFREEZE PROTEIN |
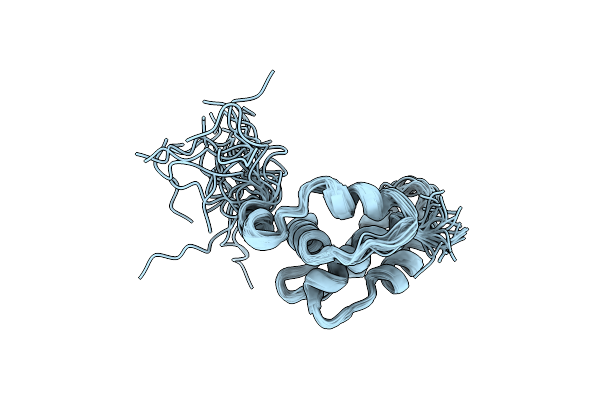 |
Organism: Tenebrio molitor
Method: SOLUTION NMR Release Date: 1999-11-10 Classification: ANTIFREEZE PROTEIN |
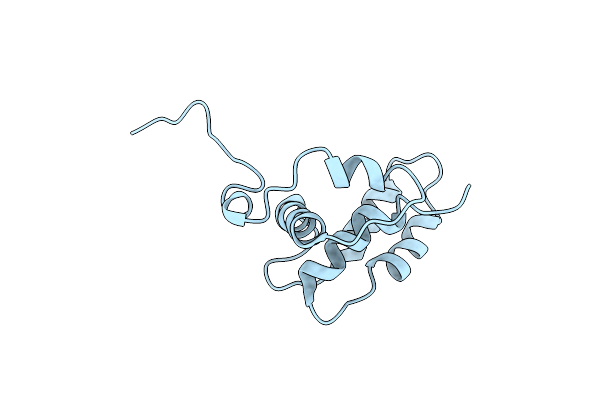 |
Organism: Tenebrio molitor
Method: SOLUTION NMR Release Date: 1999-11-10 Classification: ANTIFREEZE PROTEIN |
 |
 |
 |
 |