Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
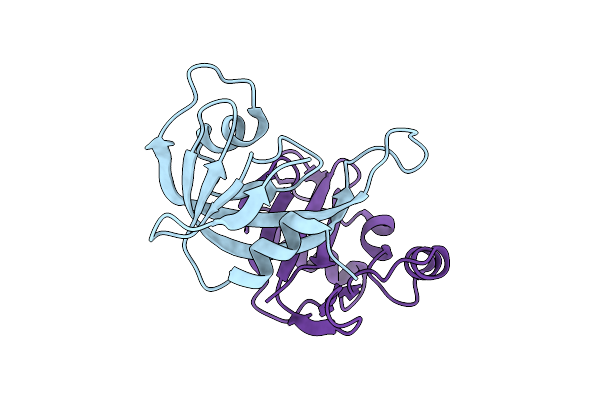 |
Crystal Structure For Yxid-Yxxd Toxin-Immunity Protein Complex From Bacillus Subtilis 6633.
Organism: Bacillus spizizenii atcc 6633 = jcm 2499
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TOXIN |
 |
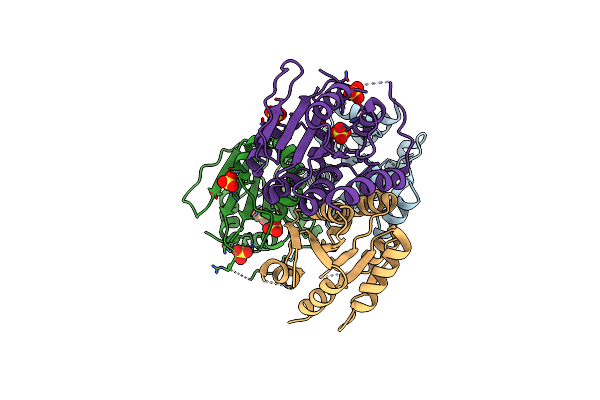
Cryo-Em Structure Of A Type Ii-D Crispr-Cas9 In Complex With Single-Guided Rna And Double-Stranded Dna
Organism: Nitrospirae bacterium rbg_13_39_12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Siamese algae-eater influenza-like virus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: HYDROLASE Ligands: NAG |
 |
Crystal Structure Of Phosphonopyruvate Decarboxylase Rhief From Bacillus Subtilis Atcc6633
Organism: Bacillus spizizenii atcc 6633 = jcm 2499
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2024-12-18 Classification: LYASE Ligands: SO4 |
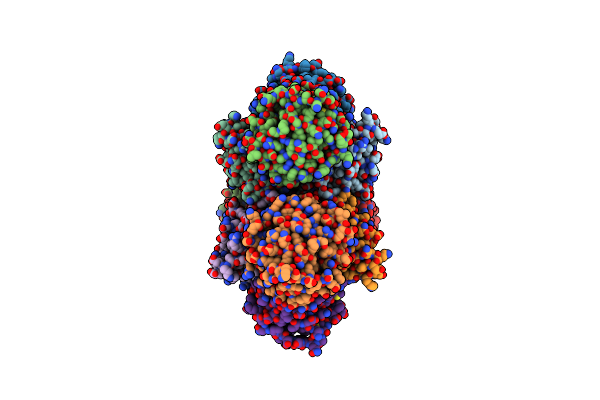 |
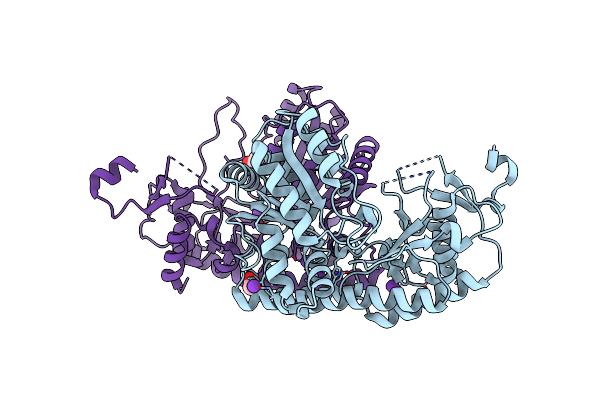
Crystal Structure Of Phosphonopyruvate Decarboxylase Rhief From Bacillus Subtilis Atcc6633 In Complex With Thiamine Pyrophosphate
Organism: Bacillus spizizenii atcc 6633 = jcm 2499
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2024-12-18 Classification: LYASE Ligands: MG, TPP, SPV |
 |
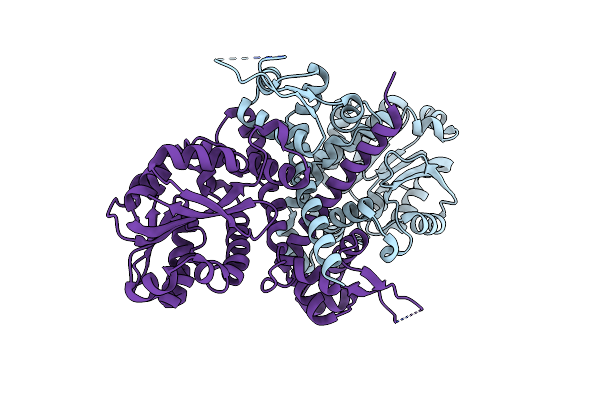
Structure Of The Thiocysteine Lyase (Sh) Domain From Guangnanmycin A Biosynthetic Pathway
Organism: Streptomyces sp. cb01883
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-09-25 Classification: BIOSYNTHETIC PROTEIN Ligands: K, ACT |
 |
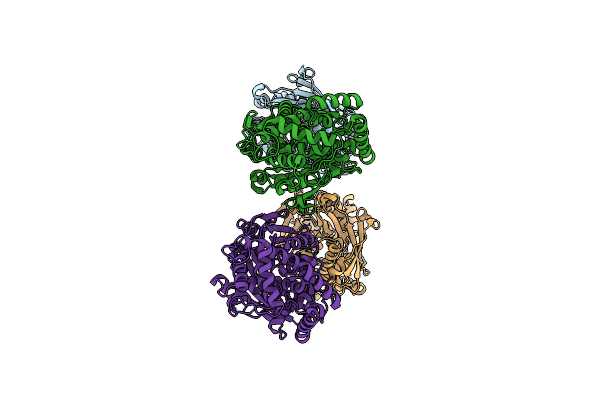
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-18 Classification: BIOSYNTHETIC PROTEIN |
 |
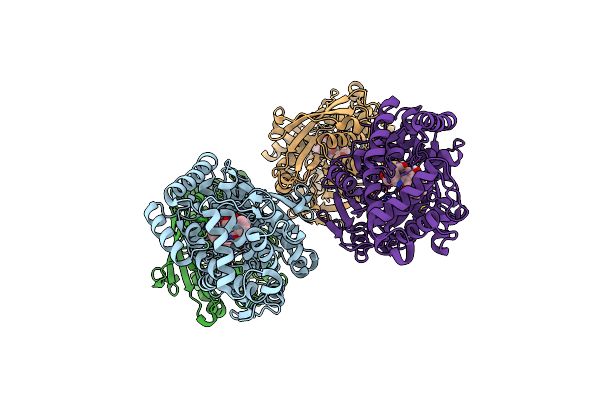
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-10-18 Classification: BIOSYNTHETIC PROTEIN |
 |
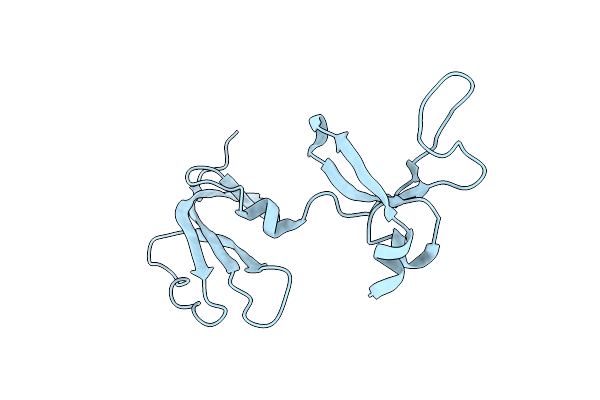
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-10-18 Classification: BIOSYNTHETIC PROTEIN Ligands: 9VG |
 |
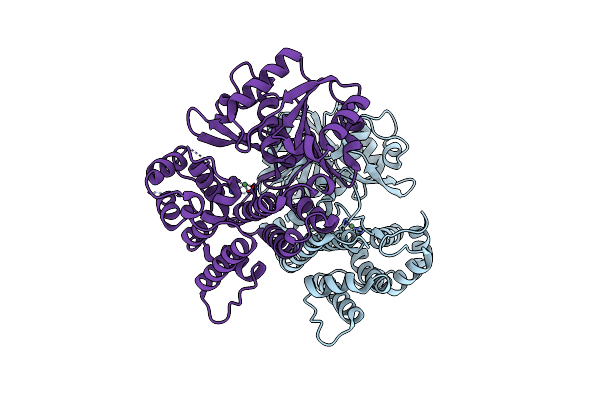
Crystal Structure Of Pilin-Specific Sortase Srtc2 From Lactobacillus Rhamnosus Gg
Organism: Lacticaseibacillus rhamnosus gg
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2023-08-30 Classification: HYDROLASE |
 |
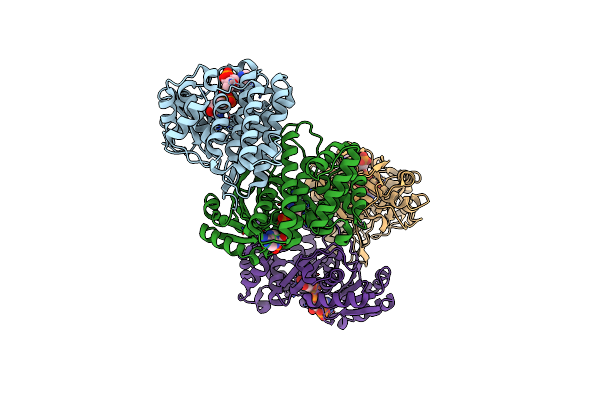
Organism: Bacillus subtilis subsp. spizizenii atcc 6633
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: NI |
 |
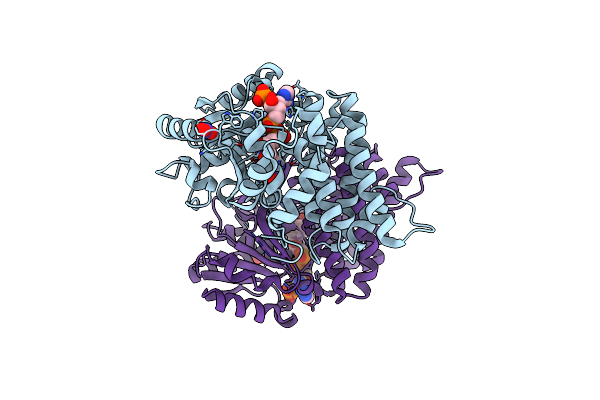
Crystal Structure Of Bacillus Subtilis Yugj In Complex With Nadp And Nickel
Organism: Bacillus subtilis subsp. spizizenii atcc 6633
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: NAP, NI |
 |
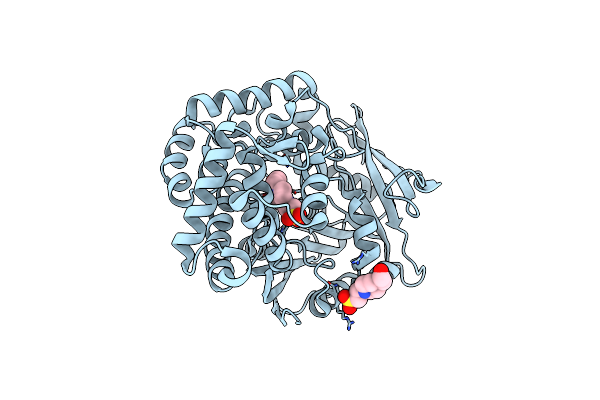
Crystal Structure Of Bacillus Subtilis Yugj In Complex With Nadp And Nitrate
Organism: Bacillus subtilis subsp. spizizenii atcc 6633
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: NAP, NO3 |
 |
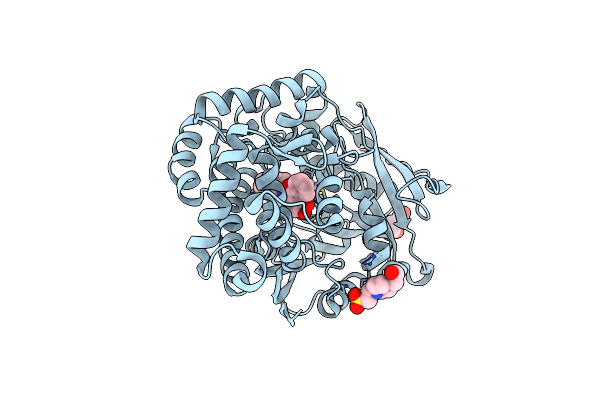
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: EPE |
 |
Organism: Streptomyces sp. cb03234
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2022-11-09 Classification: HYDROLASE Ligands: U8L, EPE, SIN |
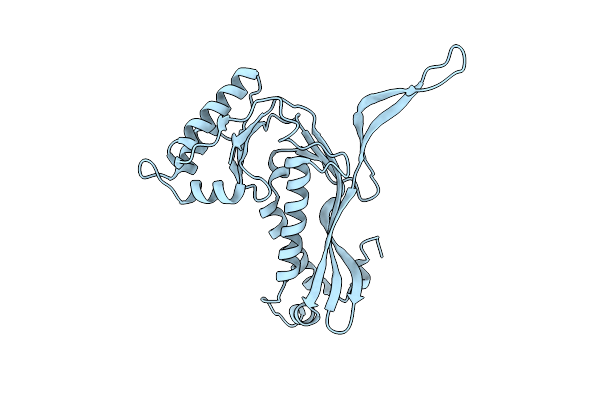 |
Organism: Brevibacterium linens
Method: ELECTRON MICROSCOPY Release Date: 2022-10-05 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacillus thuringiensis serovar andalousiensis bgsc 4aw1
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2022-04-13 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, U2F |
 |
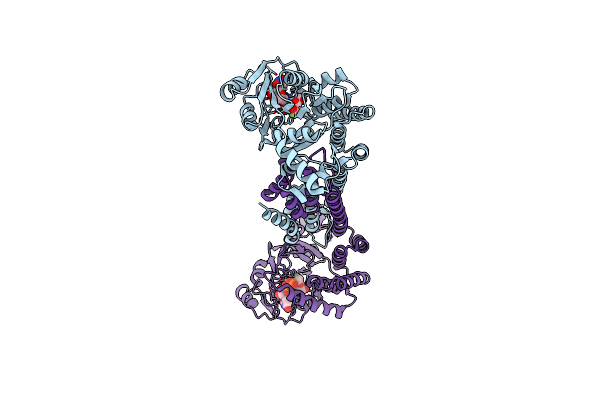
Crystal Structure Of N2, A Member Of 4-Oxalocrotonate Tautomerase (4-Ot) Family
Organism: Gammaproteobacteria bacterium sg8_31
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-06-02 Classification: ISOMERASE |
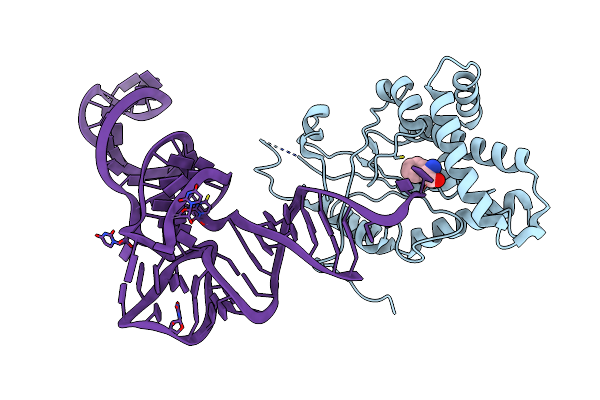 |
Structure Of Cyclodipeptide Synthase From Candidatus Glomeribacter Gigasporarum Bound To Phe-Trnaphe
Organism: Escherichia coli, Candidatus glomeribacter gigasporarum beg34
Method: X-RAY DIFFRACTION Resolution:5.00 Å Release Date: 2020-12-30 Classification: RNA BINDING PROTEIN Ligands: PHE |
 |
Crystal Structure Of Tetra-Tandem Repeat In Extending Rtx Adhesin From Aeromonas Hydrophila
Organism: Aeromonas hydrophila subsp. hydrophila (strain atcc 7966 / dsm 30187 / jcm 1027 / kctc 2358 / ncimb 9240)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-10-14 Classification: METAL BINDING PROTEIN Ligands: GOL, CL, NA, CA, PEG |