Search Count: 45
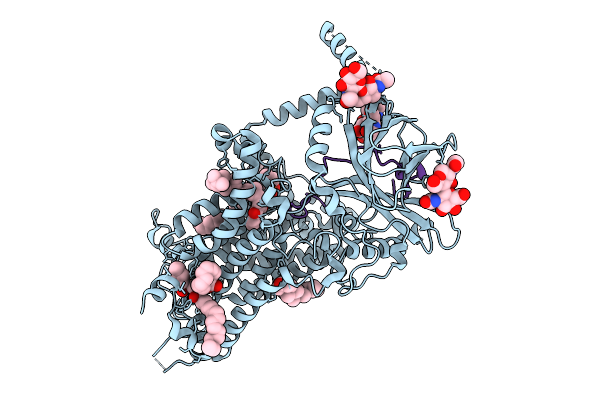 |
Vitamin K-Dependent Gamma-Carboxylase In Complex With Coagulation Factor Ix And Vitamin K
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: MEMBRANE PROTEIN Ligands: 6PL, CLR, 1L3 |
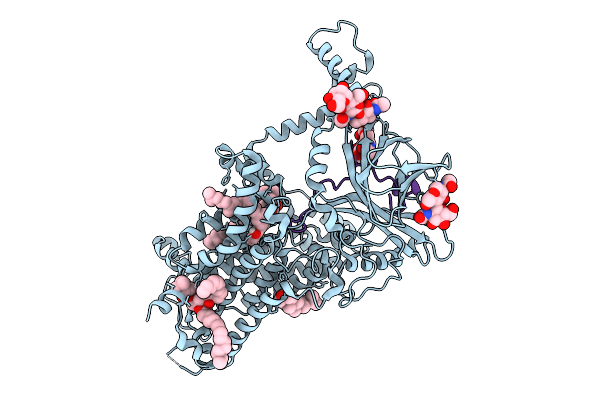 |
Double-Mutant (K217A & K218A) Vitamin K-Dependent Gamma-Carboxylase In Complex With Coagulation Factor Ix And Vitamin K
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: MEMBRANE PROTEIN Ligands: A1EMC, 6PL, CLR |
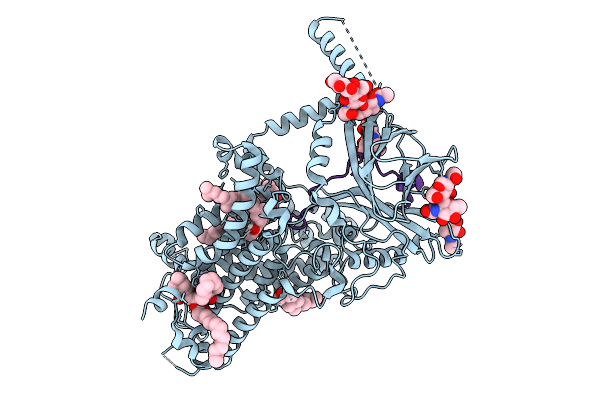 |
Double-Mutant (K217A & K218A) Vitamin K-Dependent Gamma-Carboxylase In Complex With Coagulation Factors X And Vitamin K
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: MEMBRANE PROTEIN Ligands: NAG, A1EMC, 6PL, CLR |
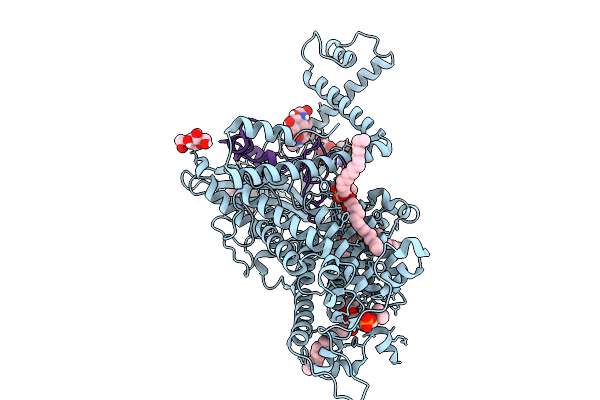 |
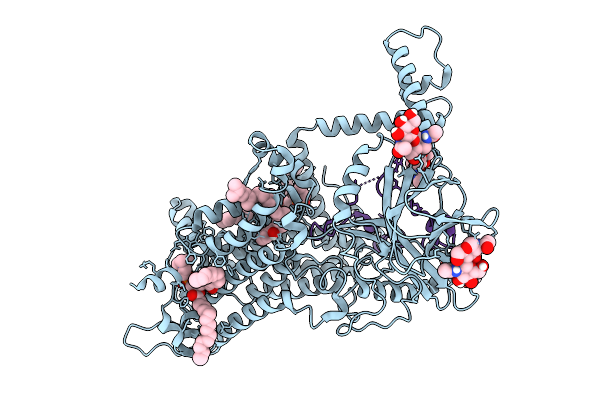
Vitamin K-Dependent Gamma-Carboxylase With Osteocalcin (Mutant) And Vitamin K Hydroquinone And Calcium
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: NAG, 6PL, A1AVC |
 |
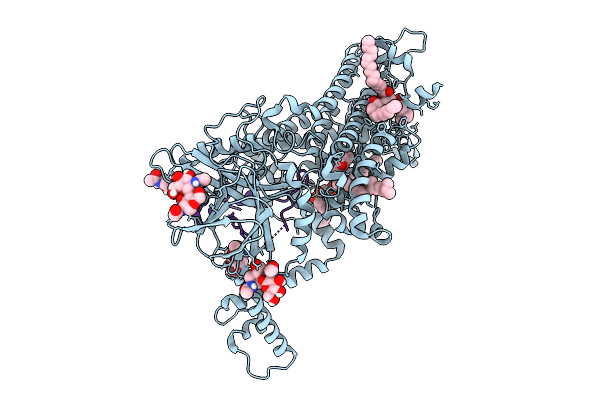
Vitamin K-Dependent Gamma-Carboxylase With Osteocalcin (Mutant) And Vitamin K Hydroquinone
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: NAG, 6PL, A1AVC |
 |
Vitamin K-Dependent Gamma-Carboxylase With Osteocalcin And Vitamin K Hydroquinone
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: TRANSFERASE Ligands: NAG, 6PL, A1AVC |
 |
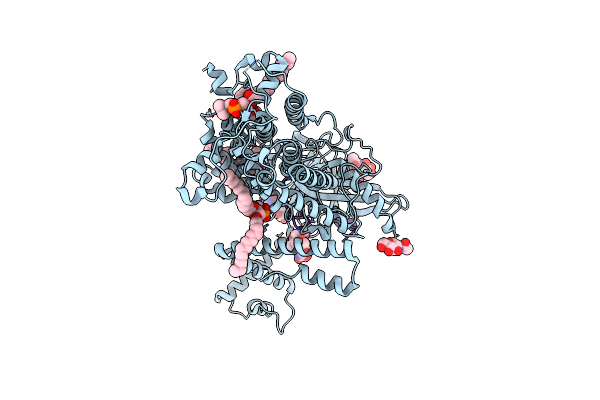
Lecithin:Cholesterol Acyltransferase Bound To Apolipoprotein A-I Dimer In Hdl
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: LIPID TRANSPORT Ligands: 6PL |
 |
Vitamin K-Dependent Gamma-Carboxylase With Factor Ix Propeptide And Glutamate-Rich Region And With Vitamin K Hydroquinone
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN,LYASE/SUBSTRATE Ligands: A1AVC, 6PL, NAG |
 |
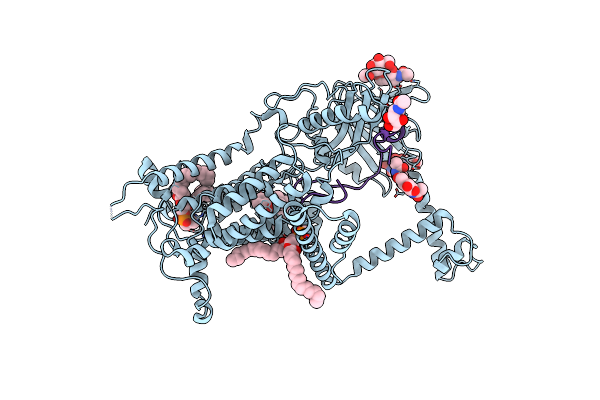
Vitamin K-Dependent Gamma-Carboxylase With Factor X Propeptide And Glutamate-Rich Region And With Vitamin K Hydroquinone
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN,LYASE/SUBSTRATE Ligands: A1AVC, 6PL, NAG |
 |
Vitamin K-Dependent Gamma-Carboxylase With Protein C Propeptide And Glutamate-Rich Region And With Vitamin K Hydroquinone
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN,LYASE/SUBSTRATE Ligands: A1AVC, 6PL, NAG |
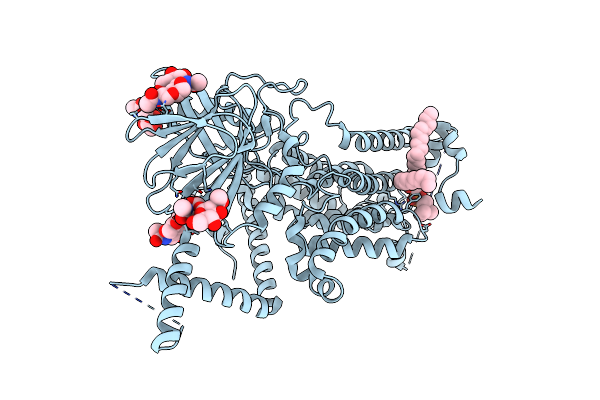 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN,LYASE Ligands: 6PL, NAG |
 |
Vitamin K-Dependent Gamma-Carboxylase With Tmg2 Propeptide And Glutamate-Rich Region And With Vitamin K Hydroquinone
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN,LYASE/SUBSTRATE Ligands: A1AVC, 6PL, NAG |
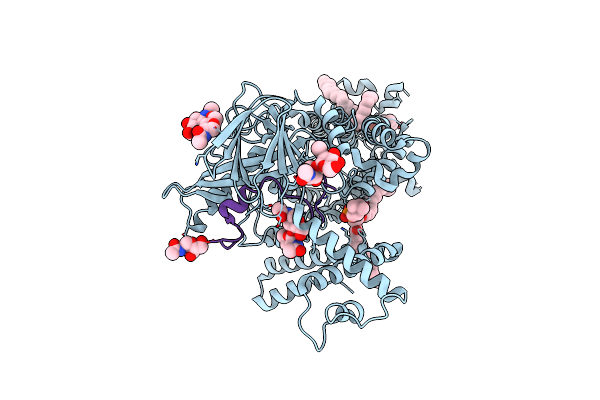 |
Vitamin K-Dependent Gamma-Carboxylase With Tmg2 Propeptide And Glutamate-Rich Region
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN,LYASE/SUBSTRATE Ligands: 6PL, NAG |
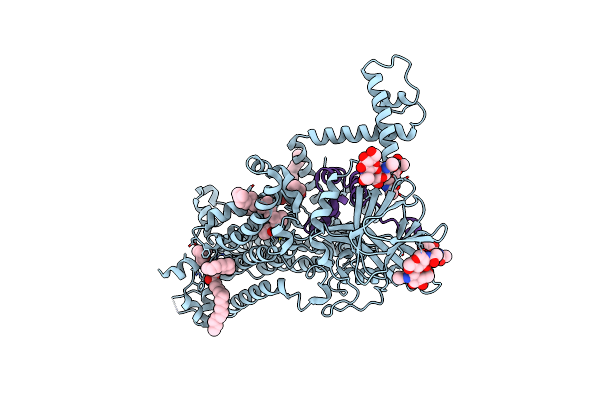 |
Vitamin K-Dependent Gamma-Carboxylase With Factor Ix Propeptide And Partially Carboxylated Glutamate-Rich Region And With Vitamin K Hydroquinone And Calcium
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN,LYASE/SUBSTRATE Ligands: A1AVC, 6PL, NAG |
 |
Homo Sapiens Xenotropic And Polytropic Retrovirus Receptor 1 (Xpr1) With Y22A/E23A/K26A Mutations
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: PO4, 6PL |
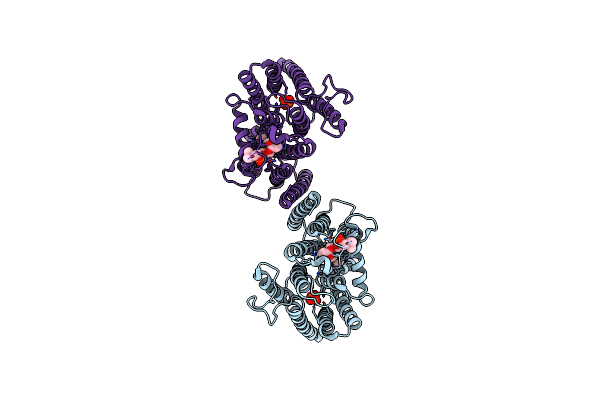 |
Wild Type Homo Sapiens Xenotropic And Polytropic Retrovirus Receptor 1 (Xpr1)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PROTEIN TRANSPORT Ligands: PO4, 6PL |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: TRANSFERASE Ligands: 6PL, A1D5V, 7PO |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: TRANSFERASE Ligands: 6PL |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: TRANSFERASE Ligands: 6PL, A1D74 |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: MEMBRANE PROTEIN Ligands: CLR, CA, 6PL, MG, K |

