Search Count: 35
 |
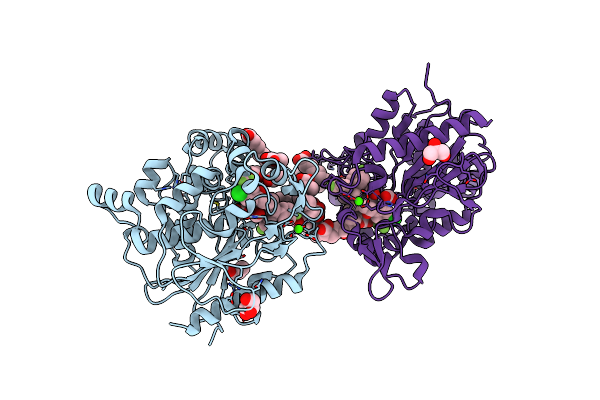
Crystal Structure Of Ubiquitin Carboxy Terminal Hydrolase L1 Q209C Mutant Covalently Crosslinked To Ubiquitin Genetically Encoded With N6-(6-Bromohexanoyl)-L-Lysine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE Ligands: 6NA |
 |
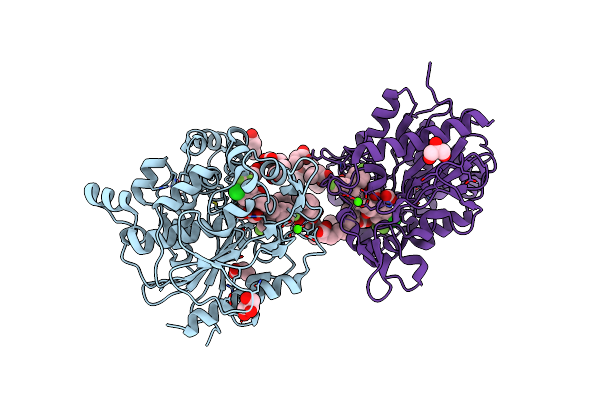 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: HYDROLASE Ligands: OCA, PPI, GOL, A1L60, FMT, ZN, CA, 6NA, CL, BUA, 11A |
 |
Organism: Actinoplanes
Method: SOLUTION NMR Release Date: 2025-04-16 Classification: ANTIBIOTIC Ligands: 6NA |
 |
Organism: Pseudomonas putida
Method: SOLUTION NMR Release Date: 2025-02-26 Classification: SURFACTANT PROTEIN Ligands: 6NA |
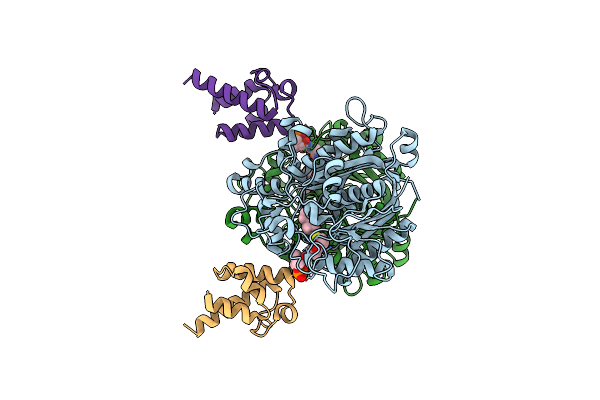 |
Crystal Structure Of Beta-Ketoacyl-Acp Synthase Fabf K336A In Complex With Hexanoyl-Acp From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-10-16 Classification: BIOSYNTHETIC PROTEIN Ligands: 6NA, PN7 |
 |
 |
 |
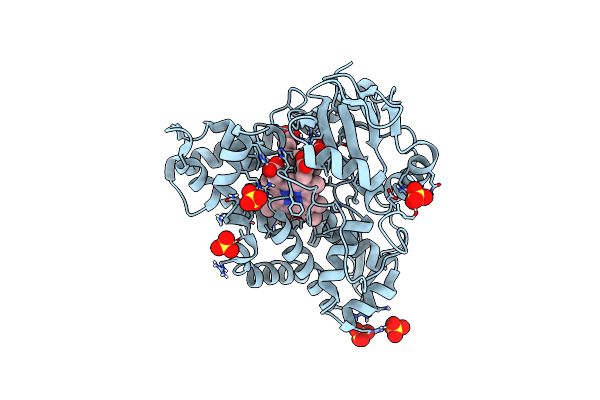 |
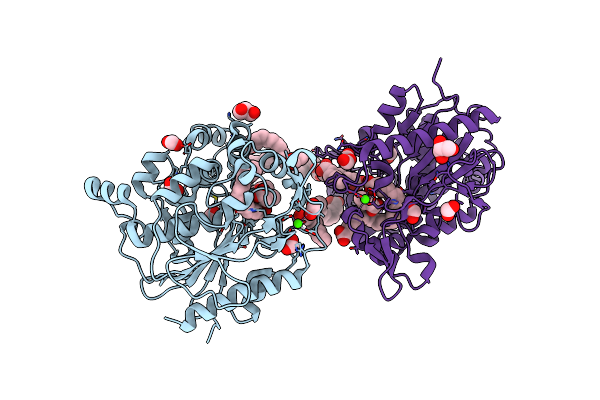
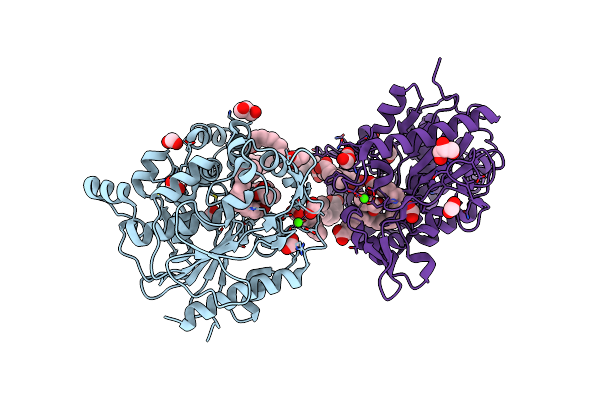
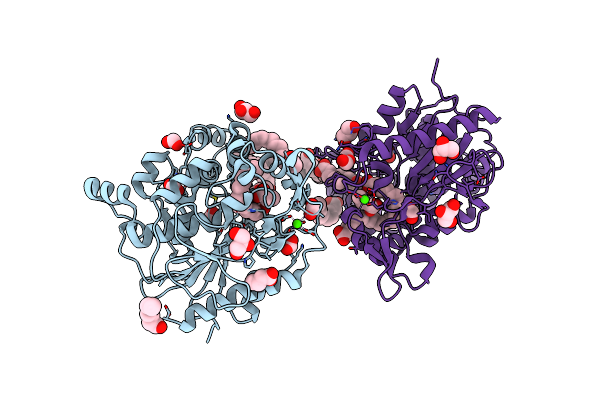
Crystal Structure Of Cyp109A2 From Bacillus Megaterium Bound With Putative Ligands Hexanoic Acid And Octanoic Acid
Organism: Priestia megaterium dsm 319
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: HEB, 6NA, OCA, SO4 |
 |
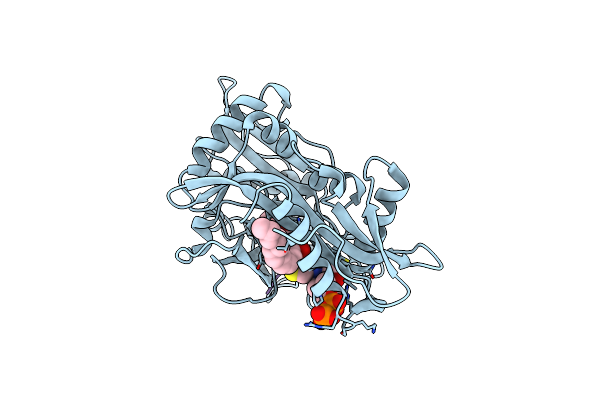
The 0.95 Angstrom X-Ray Structure Of The Human Heart Fatty Acid-Binding Protein Complexed With Hexanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2022-12-21 Classification: LIPID BINDING PROTEIN Ligands: 6NA, 1PE |
 |
Crystal Structure Of Human N-Myristoyltransferase 1 Fragment (Residues 109-496) Bound To Diacylated Human Arf6 Octapeptide And Coenzyme A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-07-27 Classification: TRANSFERASE Ligands: 4PS, MYR, 6NA |
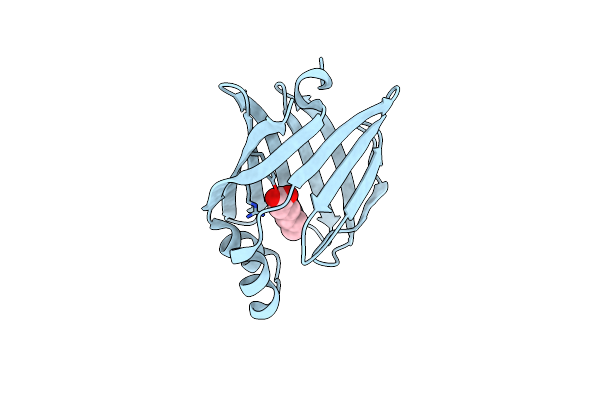 |
Room Temperature Structure Of The Human Heart Fatty Acid-Binding Protein Complexed With Hexanoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2022-07-20 Classification: LIPID BINDING PROTEIN Ligands: 6NA |
 |
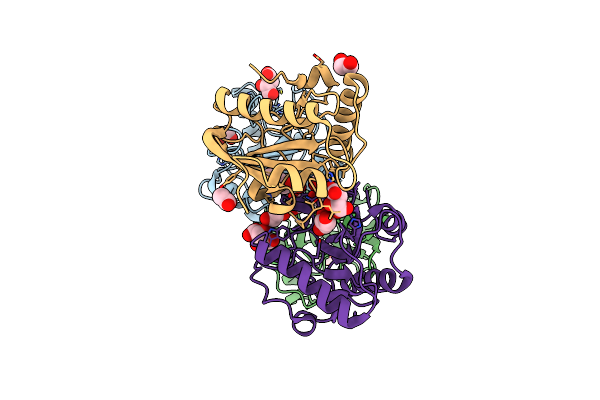
Crystal Structure Of The Ternary Complex Of Peptidoglycan Recognition Protein, Pgrp-S With Hexanoic And Tartaric Acids At 2.07 A Resolution.
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-05-11 Classification: IMMUNE SYSTEM Ligands: TLA, EDO, ACT, CL, 6NA, GOL |
 |
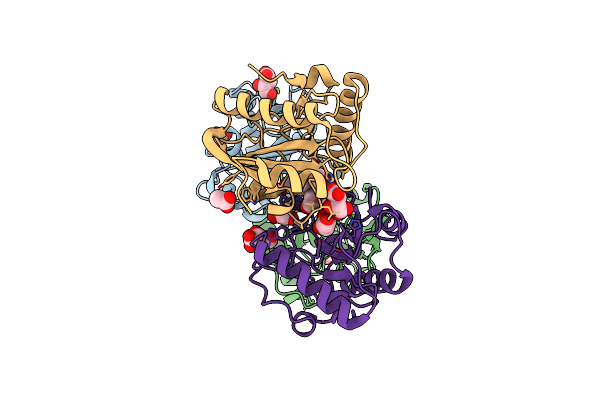
Crystal Structure Of The Ternary Complex Of Peptidoglycan Recognition Protein, Pgrp-S With Hexanoic And Tartaric Acids At 2.28 A Resolution.
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2022-05-11 Classification: IMMUNE SYSTEM Ligands: TLA, EDO, CL, 6NA, ACT, GOL |
 |
Crystal Structure Of The Ternary Complex Of Peptidoglycan Recognition Protein, Pgrp-S With Hexanoic And Tartaric Acids At 2.67 A Resolution.
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2022-05-11 Classification: IMMUNE SYSTEM Ligands: EDO, TLA, CL, ACT, 6NA, GOL |
 |
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-02-23 Classification: HYDROLASE Ligands: GOL, EDO, 6NA, DMS, CCN, D8F, SO4, DIO, BUA, NPO, NA |
 |
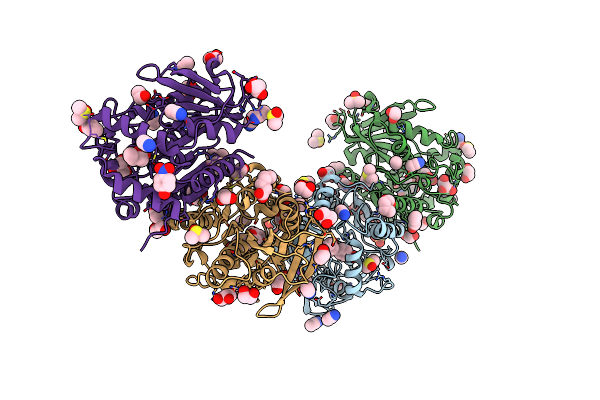
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-07-14 Classification: HYDROLASE Ligands: NPO, 6NA, PEG, EDO, DMS, DIO, GOL, SO4, PGE, NA, MES |
 |
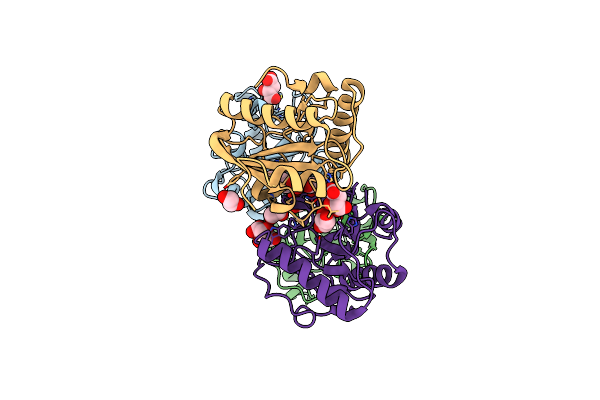
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-07-14 Classification: HYDROLASE Ligands: GOL, EDO, 6NA, DMS, CCN, D8F, SO4, DIO, BUA, NPO, NA |
 |
Structure Of The Ternary Complex Of Peptidoglycan Recognition Protein-Short (Pgrp-S) With Hexanoic Acid And Tartaric Acid At 2.30A Resolution
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-02-17 Classification: IMMUNE SYSTEM Ligands: TLA, EDO, CL, 6NA, GOL |

