Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
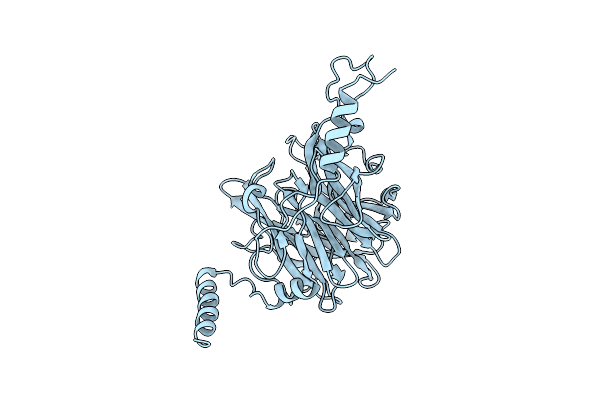 |
Crystal Structure Of Ruminococcus Albus 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase (Ramp1)
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-03-23 Classification: TRANSFERASE |
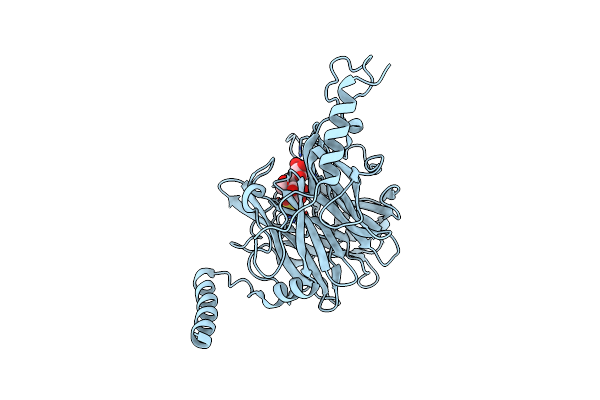 |
Crystal Structure Of Ruminococcus Albus 4-O-Beta-D-Mannosyl-D-Glucose Phosphorylase (Ramp1) In Complexes With Sulfate And 4-O-Beta-D-Mannosyl-D-Glucose
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: SO4 |
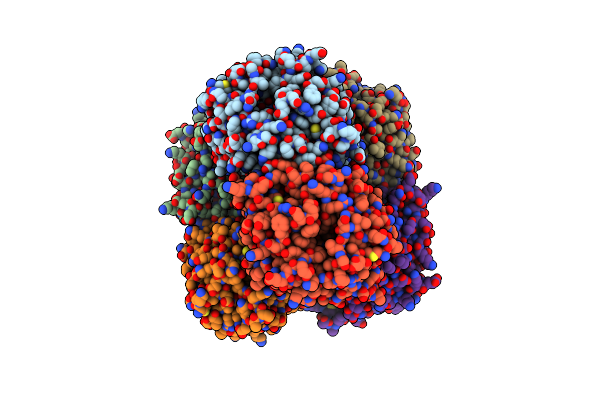 |
Crystal Structure Of Ruminococcus Albus Beta-(1,4)-Mannooligosaccharide Phosphorylase (Ramp2) In Complexes With Phosphate
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: PO4 |
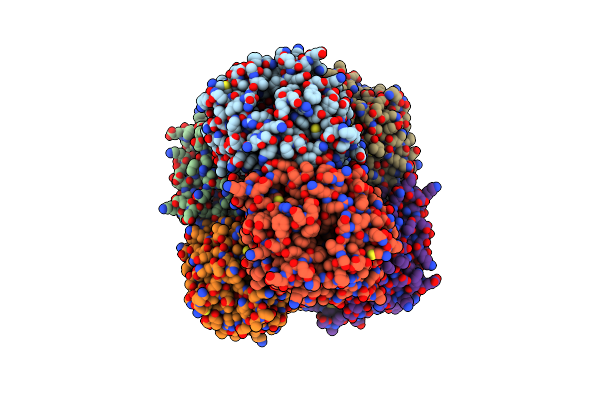 |
Crystal Structure Of Ruminococcus Albus Beta-(1,4)-Mannooligosaccharide Phosphorylase (Ramp2) In Complexes With Phosphate And Beta-(1,4)-Mannobiose
Organism: Ruminococcus albus (strain atcc 27210 / dsm 20455 / jcm 14654 / ncdo 2250 / 7)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-23 Classification: TRANSFERASE Ligands: PO4 |
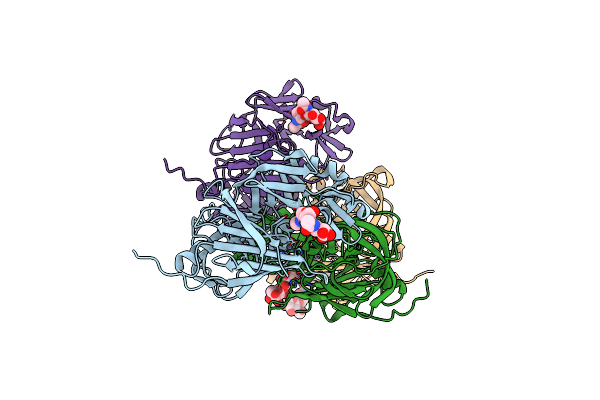 |
Organism: Blattella germanica
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2015-10-21 Classification: HYDROLASE Ligands: ZN |
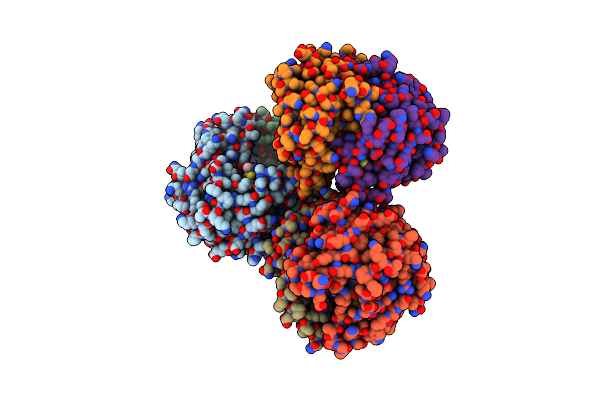 |
Organism: Blattella germanica
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-04-01 Classification: TRANSFERASE Ligands: GSH, GOL, CL |
 |
Organism: Blattella germanica
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-05-21 Classification: PROTEIN BINDING Ligands: CIT, AEF, GOL |
 |
Organism: Blattella germanica
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-05-21 Classification: ALLERGEN Ligands: CIT, GOL |
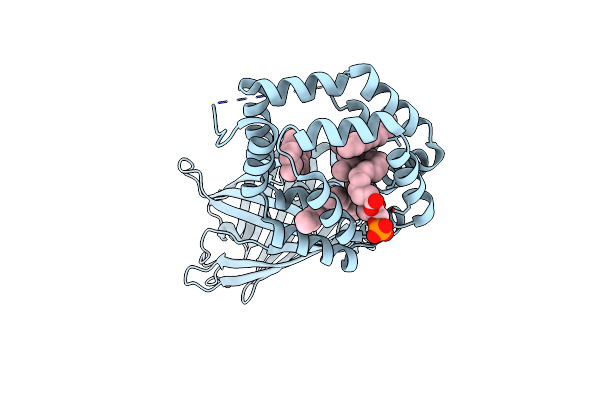 |
Organism: Aequorea victoria, Blattella germanica
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2013-07-24 Classification: LIPID BINDING PROTEIN Ligands: CL, PGT, D12 |
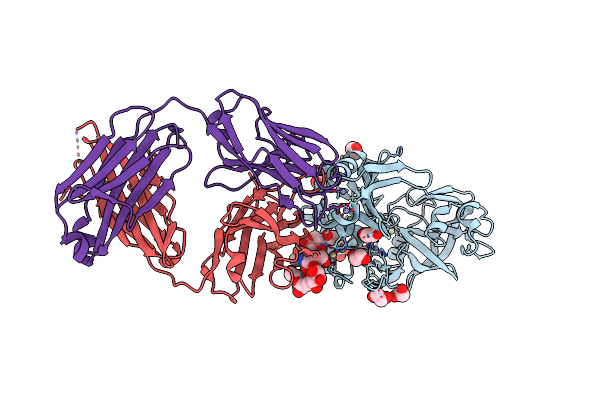 |
Organism: Blattella germanica, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-12-15 Classification: hydrolase/immune system Ligands: NAG, CD, ZN, EDO |
 |
Organism: Blattella germanica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-12-02 Classification: ALLERGEN |
 |
Organism: Blattella germanica, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2008-02-19 Classification: HYDROLASE/IMMUNE SYSTEM Ligands: NAG, ZN |
 |
Organism: Blattella germanica
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2005-03-22 Classification: HYDROLASE, ALLERGEN Ligands: NAG, ZN |
 |
 |
 |
 |
 |
 |
 |