Search Count: 2,204
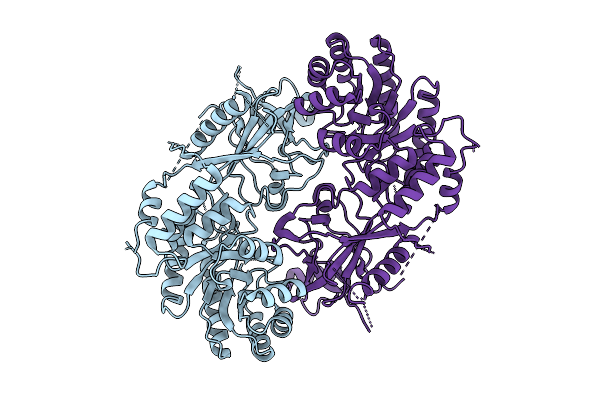 |
The Structure Of Ornithine Decarboxylase From Leishmania Infantum In Complex With Plp
Organism: Leishmania infantum
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: LYASE |
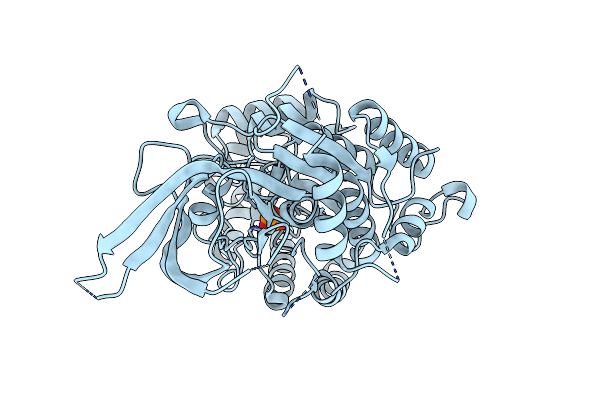 |
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LYASE Ligands: PO4 |
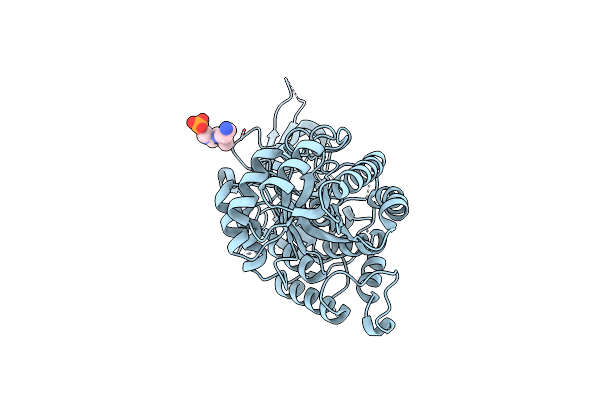 |
The Structure Of Ornithine Decarboxylase From Leishmania Infantum In Complex With Plp And Dfmo
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: LYASE Ligands: PLP, DMO |
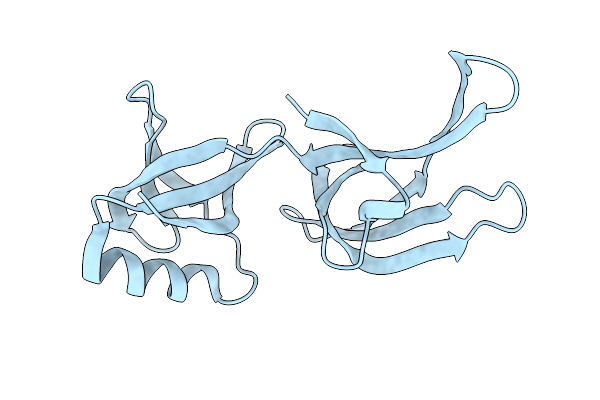 |
Hex-1 (In Cellulo, In Situ) Crystallized And Diffracted In High Five Cells. Growth And Sx Data Collection At 296 K On Crystaldirect Plates
Organism: Neurospora crassa or74a
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-01-17 Classification: STRUCTURAL PROTEIN |
 |
Organism: Larkinella arboricola
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2024-01-03 Classification: SIGNALING PROTEIN Ligands: C2E |
 |
Crystal Structure Of Acetyl-Coenzyme A Synthetase From Leishmania Infantum (Ethyl Amp Bound)
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-09-13 Classification: LIGASE Ligands: PEG, WTA |
 |
Crystal Structure Of Acetyl-Coenzyme A Synthetase From Leishmania Infantum (Ethyl Amp Bound, P21 Form)
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2023-09-13 Classification: LIGASE Ligands: CAC, GOL, CL, WTA |
 |
Crystal Structure Of Acetyl-Coenzyme A Synthetase From Leishmania Infantum (Coa And Amp Bound)
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-09-13 Classification: LIGASE Ligands: SO4, CL, AMP, COA |
 |
Crystal Structure Of Acetyl-Coenzyme A Synthetase From Leishmania Infantum (Coa, Amp And Potassium Bound)
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-09-13 Classification: LIGASE Ligands: K, CL, GOL, AMP, COA |
 |
Organism: Avian adeno-associated virus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRUS |
 |
Organism: Avian adeno-associated virus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRUS Ligands: D5M |
 |
The Crystal Structure Of Pet46, A Petase Enzyme From Candidatus Bathyarchaeota
Organism: Candidatus bathyarchaeota archaeon
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-08-23 Classification: HYDROLASE Ligands: EDO, PO4, CL |
 |
Organism: Leptospira interrogans serovar copenhageni str. fiocruz l1-130
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2023-08-23 Classification: HYDROLASE Ligands: PEG, GOL, PO4, NAD |
 |
Organism: Neurospora crassa (strain atcc 24698 / 74-or23-1a / cbs 708.71 / dsm 1257 / fgsc 987)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: FE2, CL |
 |
Organism: Neurospora crassa (strain atcc 24698 / 74-or23-1a / cbs 708.71 / dsm 1257 / fgsc 987)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: FE2, CL |
 |
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2023-05-31 Classification: UNKNOWN FUNCTION Ligands: CL, GOL |
 |
Crystal Structure Of Acetyl-Coenzyme A Synthetase From Leishmania Infantum (Amp, Acetate And Coa Bound)
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-04-19 Classification: LIGASE Ligands: AMP, ACT, COA |
 |
Structural And Biochemical Characterization Of L. Interrogans Lsa45 Reveals A Penicillin-Binding Protein With Esterase Activity
Organism: Leptospira interrogans serovar copenhageni
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-01-11 Classification: HYDROLASE Ligands: PEG, IOD, EDO |
 |
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-11-09 Classification: TRANSFERASE/INHIBITOR Ligands: FB8, EDO, CAC |
 |
Organism: Leishmania infantum
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-11-09 Classification: TRANSFERASE/INHIBITOR Ligands: 8II, EDO, SO4 |

